15 Open-source Free Web-based 3D Molecular and Protein Modelling Software

What is WebGL?
Nowadays WebGL is a trending technology as it allows developers to create complex 3D interactive graphics, games using modern browsers as a client, without the need to install additional plugins, extensions, or software. WebGL allows the browser to work directly with the GPU (graphics processing unit).
There are several JavaScript frameworks aiming to ease the development and production of 3D WebGL apps, they are classified as graphics engine, game engine, and graphics libraries. There are also WebGL-based frameworks built for Virtual Reality (VR) & Mixed Reality (MR).
WebGL for scientific and medical apps
It's a progressing technology, it's used now to create 3D web-based games, interactive 3D showcase, and simulations. It's also used in medicine, and we have covered the usage of WebGL in medical applications like Web-based DICOM apps like Med3Web, & Anatomy apps.
Benefits of WebGL
- Runs directly from the web browser
- Supported by modern web browsers
- Works out of the box does not require plugins, browser extensions, or additional software
- Smooth performance
- Embeddable with Web pages
- Allows building apps with remote collaboration
WebGL technology has been used to power several 3D molecules, protein, & DNA/ RNA rendering. In this article, we will cover the open-source projects that aim for researchers and developers.
Open source Web-based 3D Molecular & Protein Modelling software
1- LiteMol: 3D macromolecular

LiteMol is a 3D macromolecular renderer, It's built as a web & browser-based app, it comes with a set of open source tools to build heavy-duty libraries and apps like 3D molecular data streaming, & 3D volumetric data streaming.
Highlights
- Works smoothly with modern browsers: Google Chrome, Mozilla Firefox, & Safari.
- Simple API
- Modular architecture that supports plugins development
Features
- Standard visualizations: cartoons, surface, balls, and sticks, etc.
- Assemblies and symmetry mates.
- Electron Density and CryoEM maps.
- Integration with PDBe API: view and explore validation and annotation data.
- Integration with the Coordinate Server: download only parts of structures you are interested in.
- Support for the BinaryCIF format that reduces the amount of data that needs to be sent to the client several times.
Resources: GitHub
2- ChemDoodle
The ChemDoodle Web Components library is an open source and free HTML5 toolkit for building scientific WebGL applications. 2D components are also available, including chemical structures, sketchers, spectra, periodic tables, reactions and more!
The ChemDoodle Web Components library is an open source and free HTML5 toolkit for building scientific WebGL applications. It builds 2D and 3D still and interactive graphics with ease. It is tested on all of the major web browsers like Google Chrome, Mozilla Firefox, Apple Safari, Microsoft IE/ Edge, & Opera.
The ChemDoodle comes with a powerful 3D editor, that is optimized for desktop and mobile devices, a chemical sketcher app, and battle-tested GPL-released code.
3- NGL Viewer: WebGL Viewer for Molecular Visualization
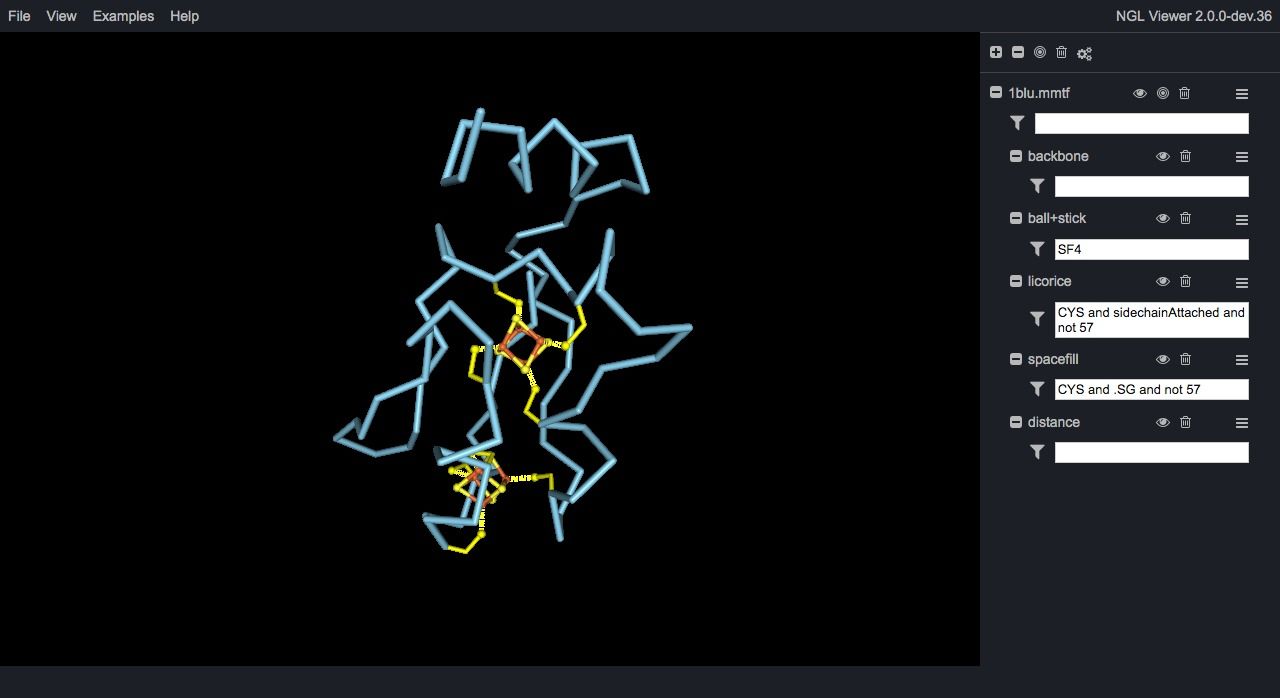
NGL Viewer is a web application for molecular visualization. WebGL is employed to display molecules like proteins and DNA/RNA with a variety of representations. Src
Ngl is an open source WebGL molecular visualization library, built to visualize proteins, & DNA/RNA structures. It provided an embeddable script for easy embeddable graphics into any HTML5-based webpage.
Ngl combines many graphics and libraries to make it easy for developers to build an interactive 3D molecular graphics, It uses Three.js the WebGL JavaScript framework for advanced 2D/ 3D graphics, among several others libraries as parsers, testing frameworks, virtual DOM manipulation libraries.
Features:
- Models, & renders molecular structures (mmCIF, PDB, PQR, GRO, SDF, MOL2, MMTF)
- Models, & renders Density volumes (MRC/MAP/CCP4, DX/DXBIN, CUBE, BRIX/DSN6, XPLOR/CNS)
- Interactive tools with the mouse, the keyboard
- Coordinate trajectories (DCD & PSF, NCTRAJ & PRMTOP, TRR/XTC & TOP, remote access via MDSrv)
4- GLMol
GLmol is an open source WebGL viewer project that allows developers and researchers to render interactive 3D molecular graphics and embed it in any webpage with ease. It's based on WebGL and JavaScript. GLmol displays biological assembly, displays unit cell, shows crystal packing and provides surface calculation and visualization. It's released under a dual license of LGPL3 and MIT license.
5- Speck

Speck is an open source 3D molecule renderer aiming to produce attractive 3D figures.
Features
- Ambient Occlusion: Ambient occlusion provides much more information about molecule structure than simple direct lighting
- Pixel-perfect atoms and bonds: Speck renders atoms and bonds not with polygons, but with imposters
- Depth-aware outlines: renders atom outlines that are depth-aware.
- Depth of field
- Mix and match rendering options
6- HTMD: Programming Environment for Molecular Discovery
HTMD is an open source Python-based programmable environment aiming to prepare, handle, simulate, visualize and analyze molecular systems. HTMD stands for (High-Throughput Molecular Dynamics). It provides easy molecule manipulation in few lines with the support of a powerful atom selection language and integrated visualization with WebGL and VMD, and comes with automated system building. HTMD comes with a comprehensive guide for developers in how to use and program it.
Highlights:
- Works with complex structure
- WebGL support
- Simple API
- Easy to learn
- Interaction toolset
7- Miew – 3D Molecular Viewer by EPAM (Med3Web)
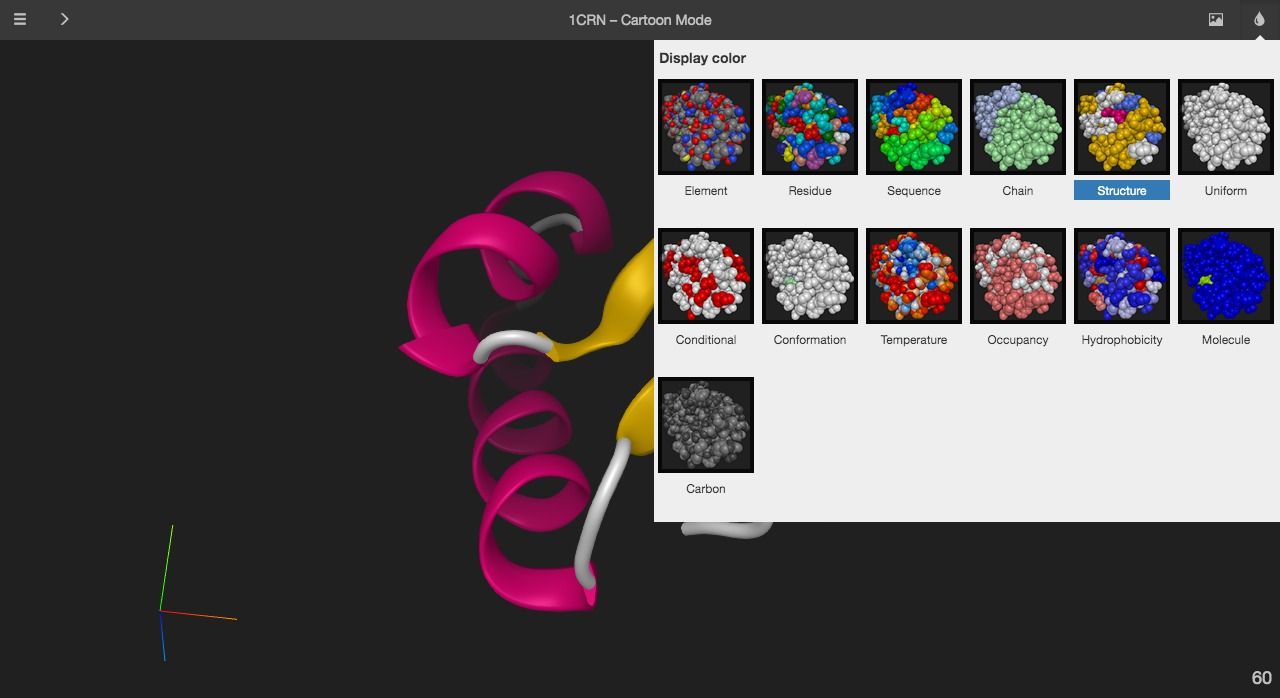
Miew is an open-source 3D molecular viewer built for advanced visualization and manipulation of molecular structures. It is created and maintained by the same company (EPAM Systems, Inc) which created Med3Web a 2D/ 3D interactive open source DICOM viewer. It can work as a standalone project or used as an integrated piece of an app
Highlights
- Works smoothly with Google Chrome, Mozilla Firefox
- Multiple display modes
- Interactive viewer

8- iView: Interactive WebGL Visualizer for Protein-ligand Complex
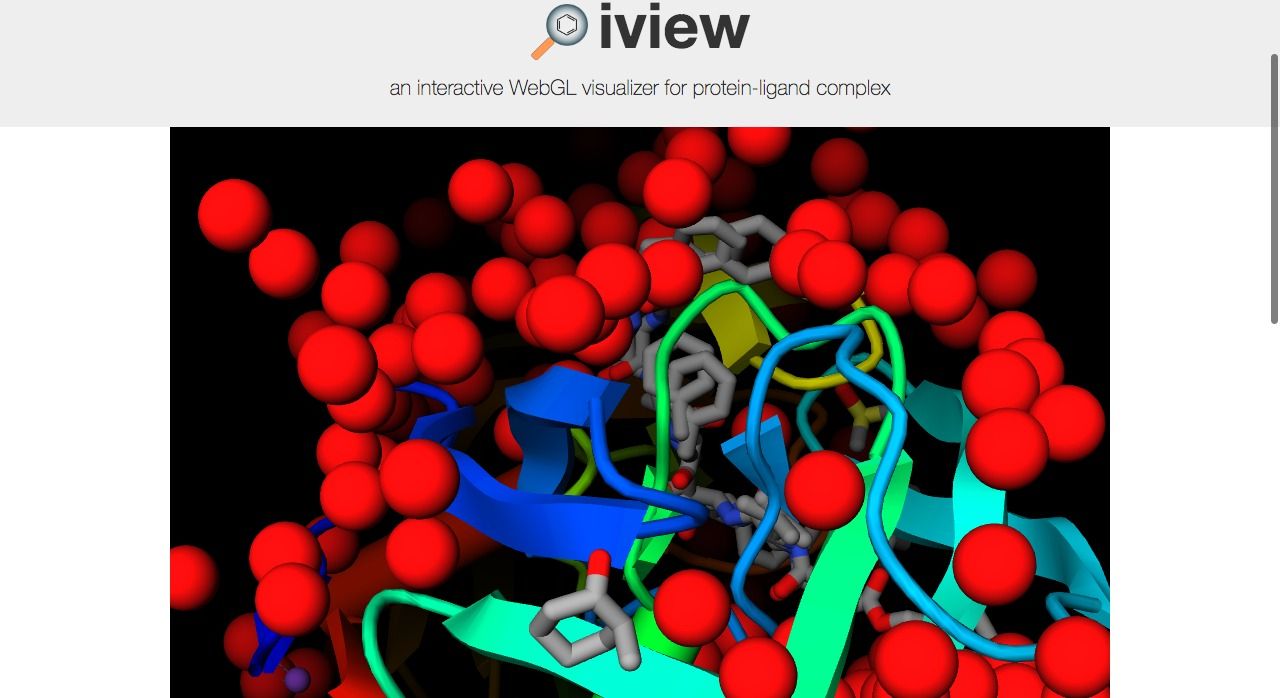
iView is an open source WebGL viewer and visualizer for protein-ligand complex, It comes with several viewing tools, and display options to display complex 3D protein molecules, It is built on top of GLmol: a 3D molecular viewer based on WebGL and Javascript, uses Three.JS 3D WebGL library, and specific libraries to parse and process molecule data.
Highlights:
- load PDB files
- Export to PNG
- Shortcuts support
- Multiple views & display options
9- NGLView
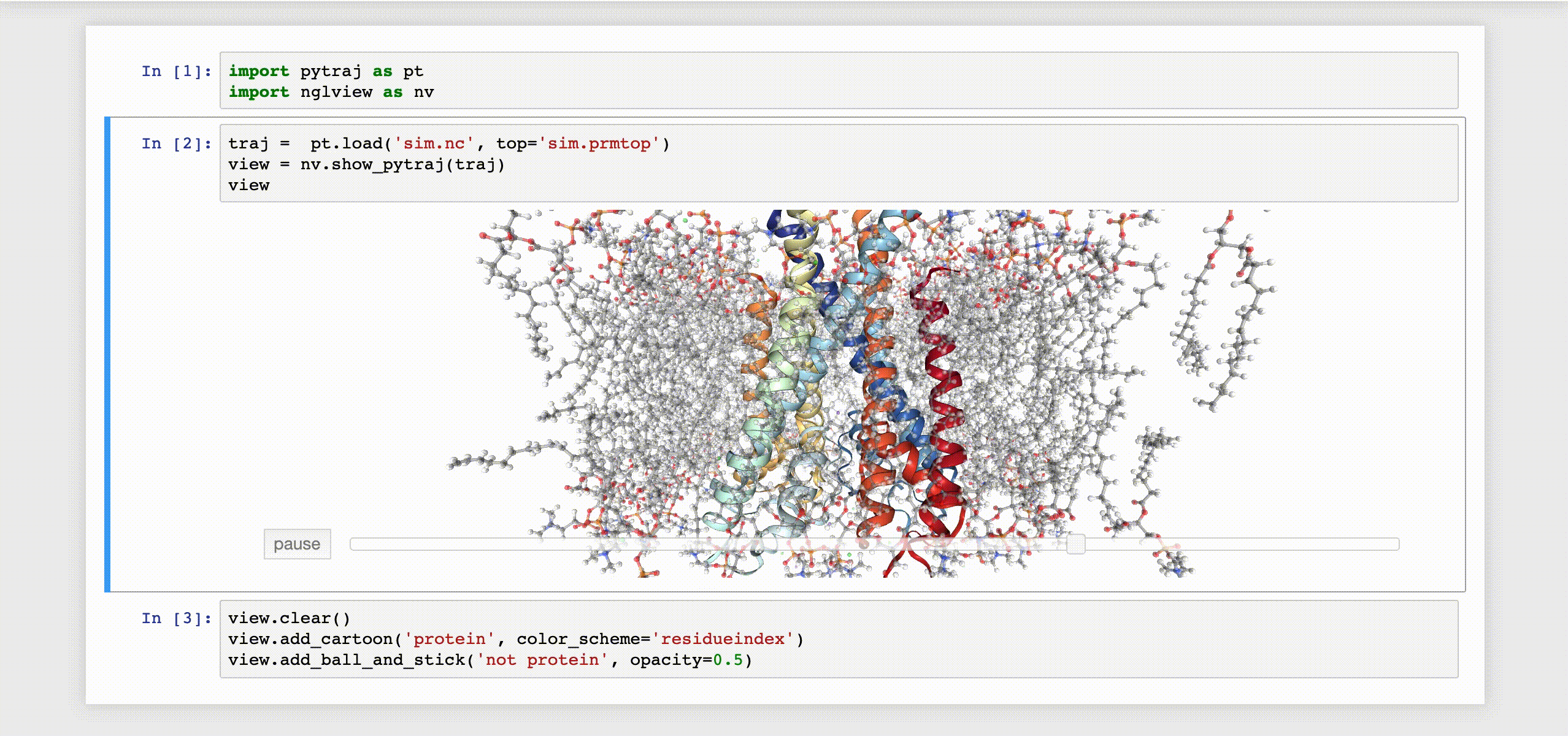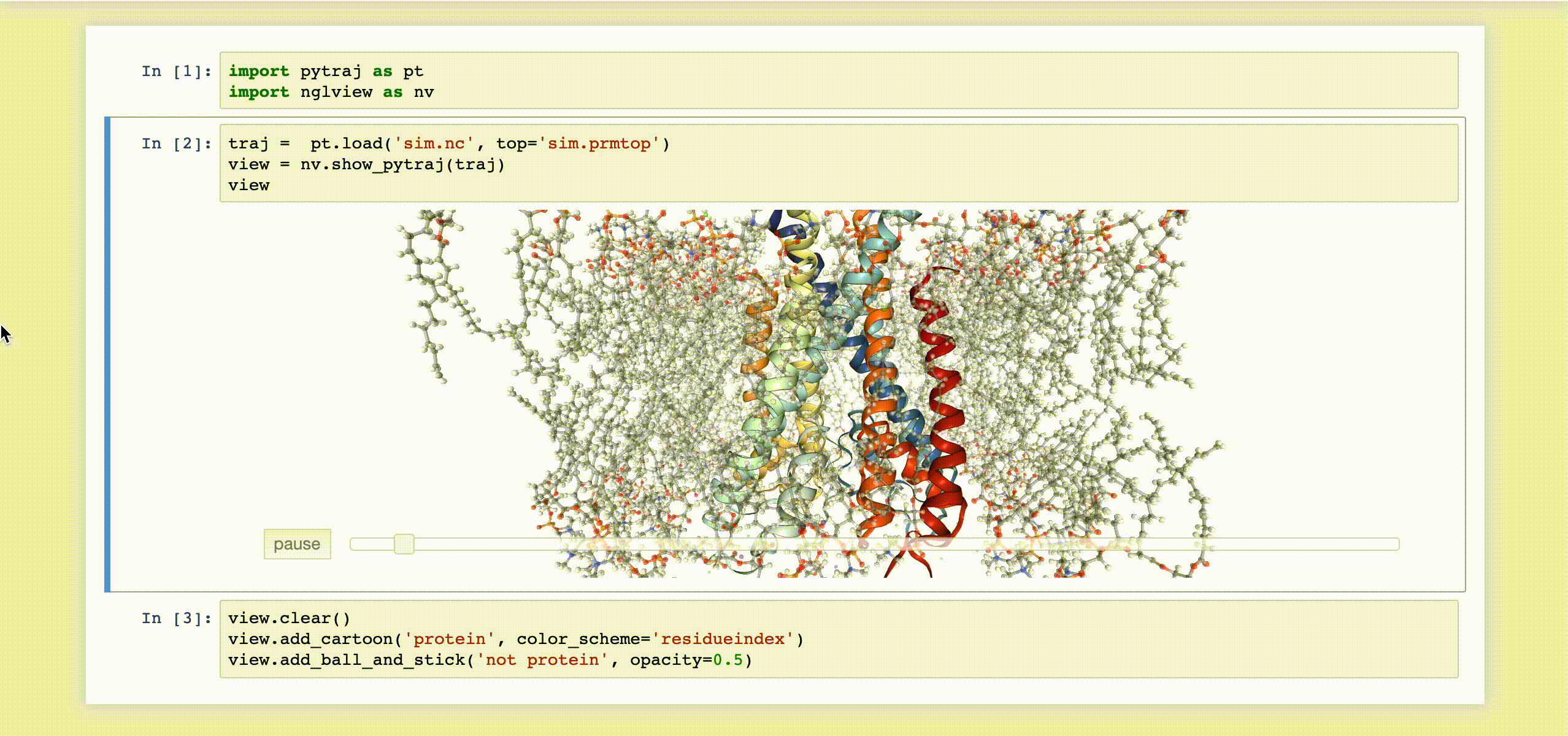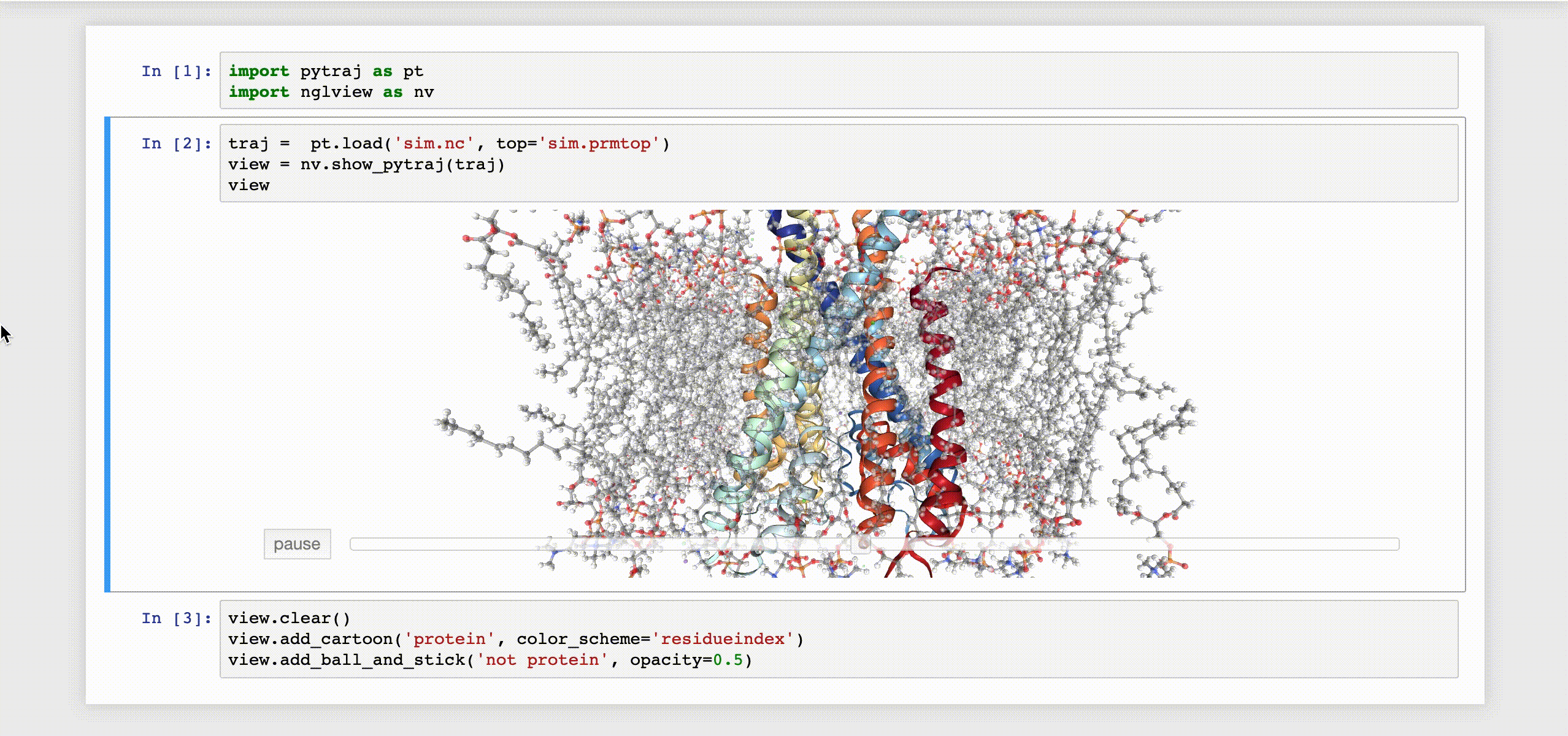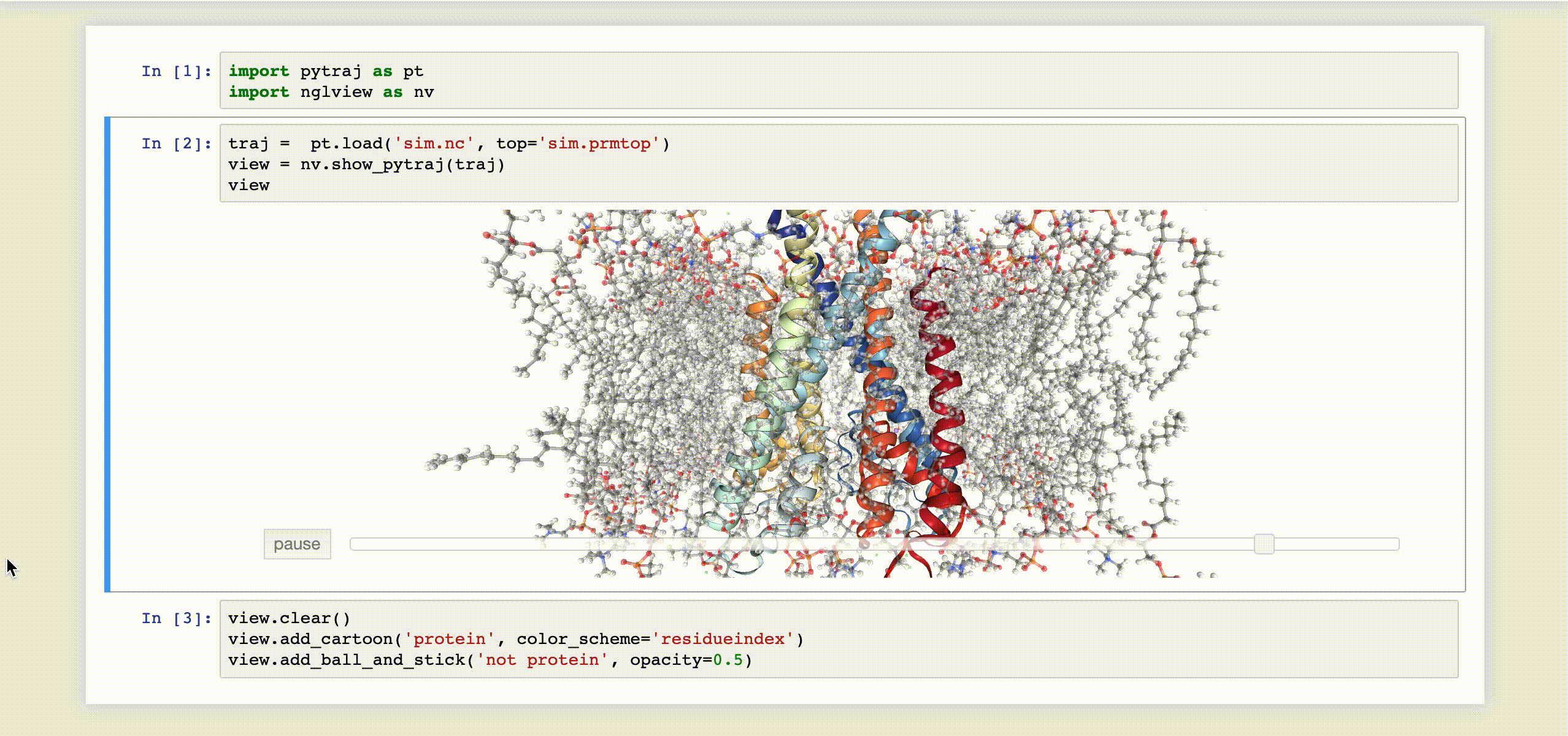
NGLView is an IPython/Jupyter interactive widget that uses NGLViewer to display 3D molecules within Jupyter.
10- MOLVWR
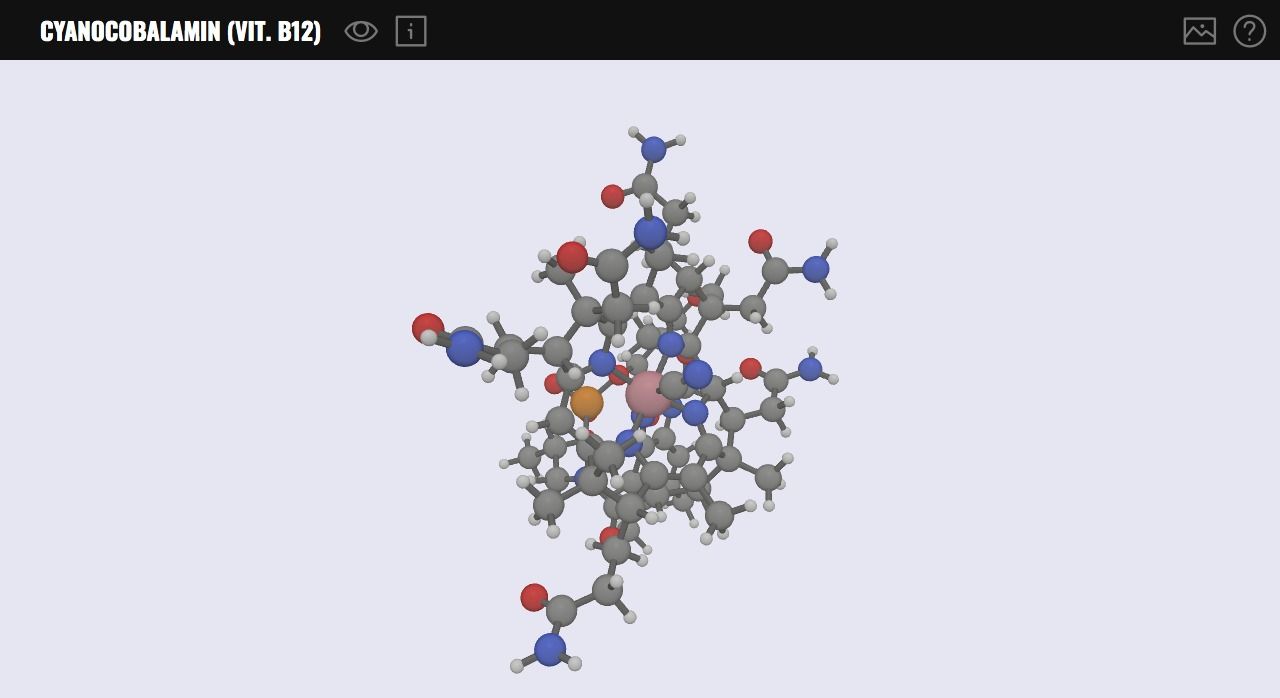
MolVWR is a 3D molecule viewer library based on Babylon.js: a WebGL JavaScript Engine. It provides a powerful layout to display and render complex 3D molecules within the browser. It has basic viewing tools, and support image exporting. It can be easily extended by JavaScript developers or integrated into existing applications or projects.
11- MolView: Web-based 3D Molecule editor
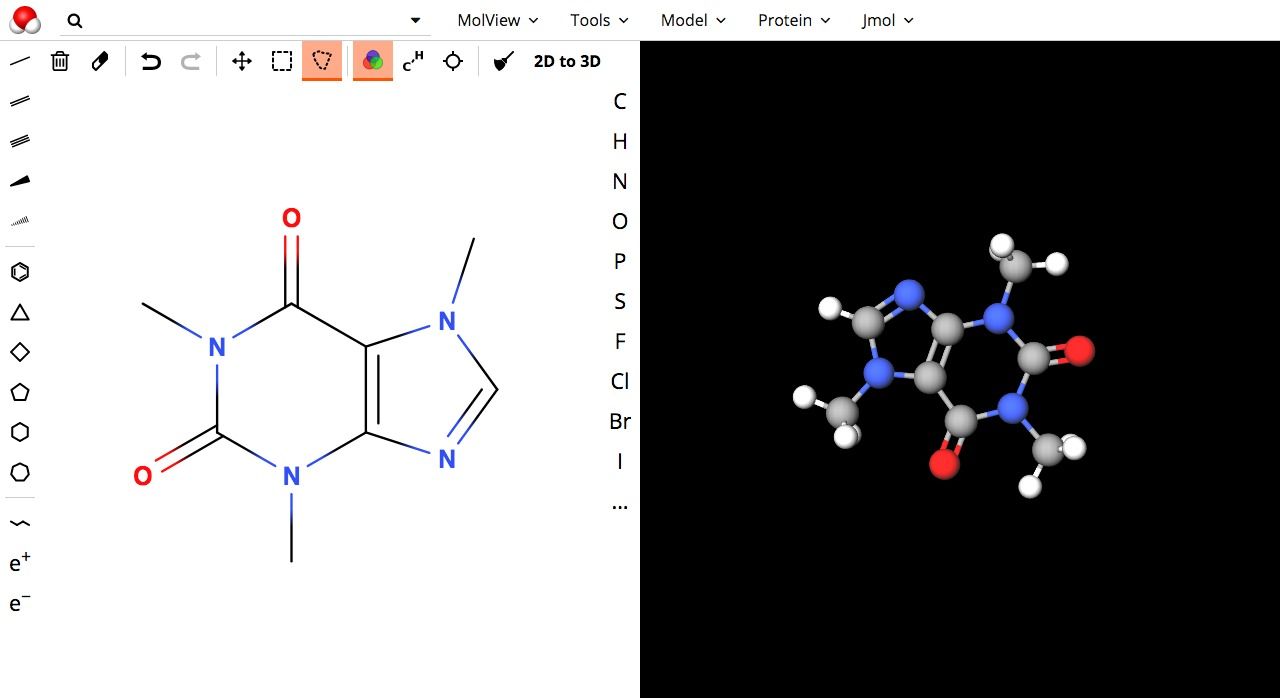
MolView is a web-based online 3D molecule editor that is powered by WebGL. It provides an editor and viewer for 2D/3D molecular structure, and advanced editor with rich features set to draw, build a molecular structure from within the browser. It supports touch devices as Android tablets and iPad.
On a technical note, MolView uses different engines and libraries and allows the user to select one of them for rendering and viewing.
Highlights
- Powerful editor
- Embedded code export
- Export still image
- Export 3D image
- Export Mol file
- Multiple engine and frameworks
12-Pymol2GlMol
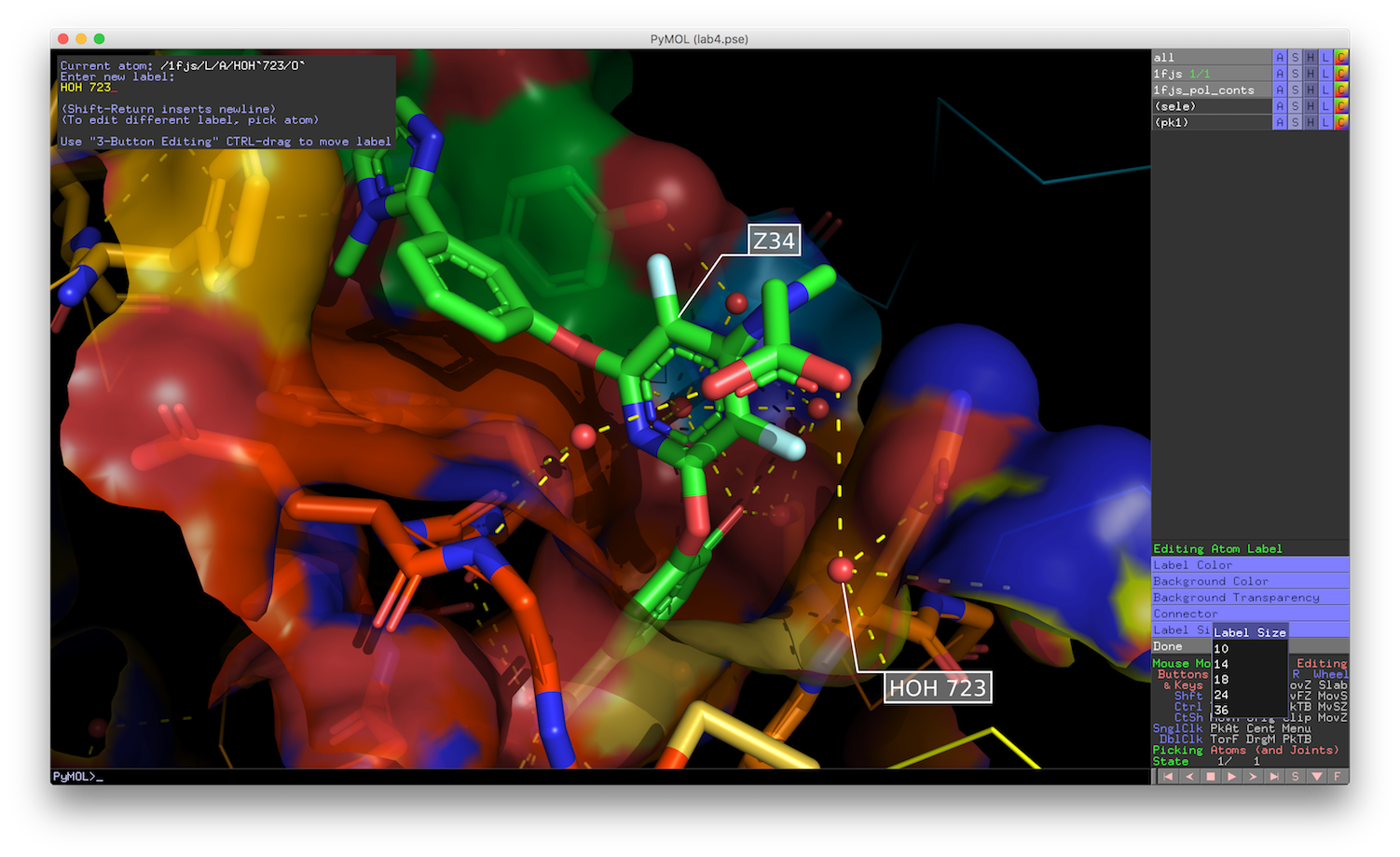
Pymol2GlMol is a script to export the 3D molecule scene from PyMOL to GLMol to work in the browser with WebGL support. PyMOL is an open-source molecular visualization system created by Warren Lyford DeLano, It's a desktop application written in Python and works on Windows, Mac OSX and Linux (Ubuntu, Debian, Fedora, LinuxMint). It provides code-based visualization for molecules.
13- Protein Viewer (PV) WebGL protein viewer
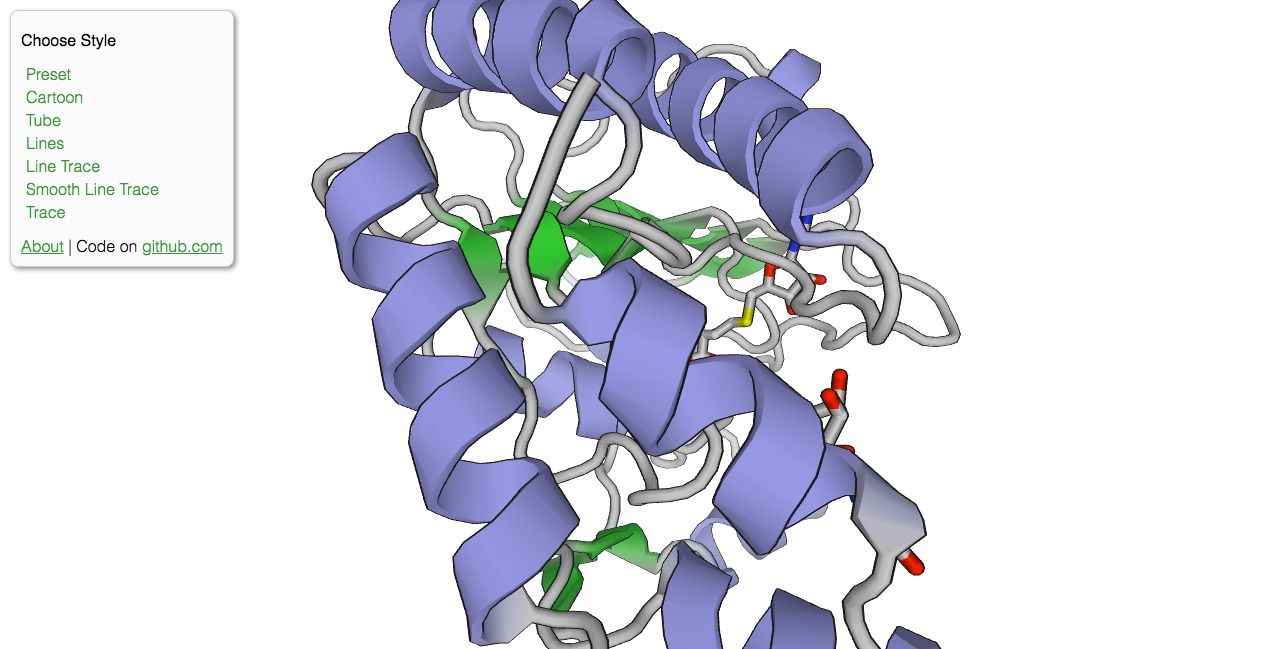
PV is an open-source protein molecule viewer that uses WebGL to render complex 3D protein structure within the browser without the need to install extra extension or plugins. Protein Viewer is based on BioJS.
The project has not been updated for years and no longer maintained, but the code is released as an open-source project.
14- BioJS
BioJS is a framework that is formed of a set of hundreds of other JavaScript libraries that gives the developers usable tools to build BioInformatics applications.
15- MDsrv: MD Trajectory Sٍerver
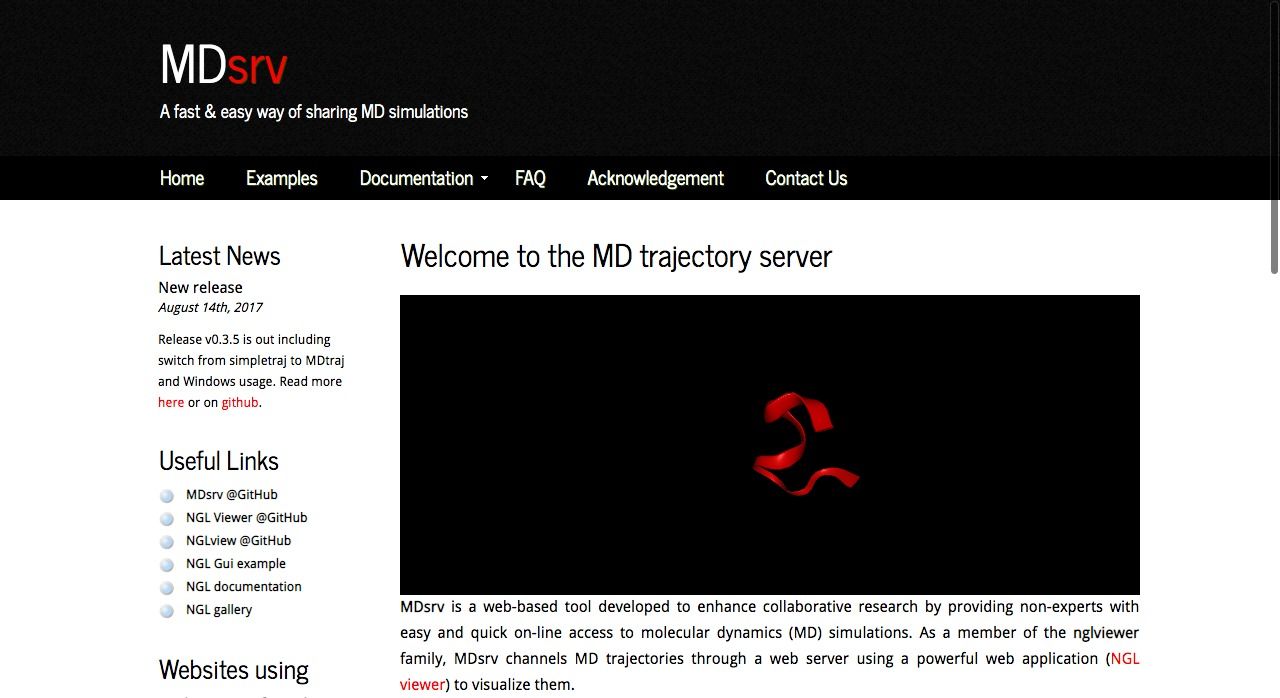
MDsrv is an open source web-based tool developed to enhance collaborative research by providing non-experts with easy and online quick access to molecular dynamics (MD) simulations, It uses WebGL to render the 3D interactive graphics in the browser. It's released under MIT license. MDsrv is packed with RESTful API which makes it a preferred option to work with remote data.
Highlights
- Direct meaningful display of raw simulation data by automatically handling periodic boundary conditions, centering, and superposition.
- Further on-the-fly MD trajectory processing (e.g. by filtering out frames and/or atoms) allows generating customized sessions that can be easily shared.
- Specific self-written functions such as 'show/hide ligand' can be added to customized sessions.
- MDsrv can display large molecular structures, densities and animate MD trajectories to allow interactive exploration and collaborative visual analysis within a local network or over the Internet.