Top 7 Linux Distros for Medical and Scientific Communities
Linux ecosystem is very big and diverse, and there are some Linux distros that aim specifically for medical and scientific communities. Although you can install specific packages in any Linux distro, a specialized distro will save time if you want to unify the working environment in labs or small institutions for example. In the following list, we take a look at the best medical and scientific Linux distros.
1- Lin4Neuro
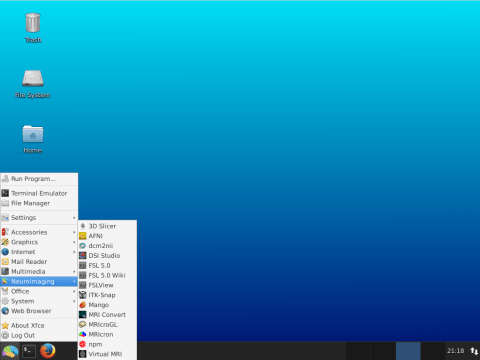
Lin4Neuro is a customized ubuntu distro especially for neuroimaging analysis. It is developed by Kiyotaka Nemoto (Associate Professor at Department of Psychiatry, Faculty of Medicine, University of Tsukuba, Japan). Lin4Neuro can be a good primer for beginners of neuroimaging analysis or students who are interested in neuroimaging analysis. It also provides a practical means of sharing analysis environments across sites.
Applications
- 3D Slicer - image analysis algorithms for diffusion tensor imaging, functional magnetic resonance imaging, and image-guided therapy.
- AFNI - for processing and displaying functional MRI data.
- UCL Camino Diffusion MRI Toolkit - for diffusion MRI processing.
- Caret - enables the user to create, view, and manipulate surface reconstructions of the cerebral and cerebellar cortex.
- Connectome Analyzer - analyze connectomes from any source (DSI, DTI, QBall, rs-fMRI, etc…) at different scales (global, subnetworks and local).
- Connectome Viewer - connects Multi-Modal Multi-Scale Neuroimaging and Network Datasets For Analysis and Visualization in Python.
- DSI Studio - for diffusion MR images analysis.
- FSL - image analysis and statistical tools for fMRI, MRI, and DTI brain imaging data.
- Ginkgo CADx - extensible multi-platform Open Source Medical Imaging software which provides a complete DICOM Viewer solution.
- ITK-Snap - segment structures in 3D medical images.
- Minc toolkit - manipulate MINC files.
- MITK - for development of interactive medical image processing software.
- MITK Diffusion - a selection of image analysis algorithms for the processing of diffusion-weighted MR images.
- MRIConvert - a medical image file conversion utility that converts DICOM files to NIfTI 1.1, Analyze 7.5, SPM99/Analyze, BrainVoyager, and MetaImage volume formats.
- MRIcron - GUI-based visualization and analysis tool for (functional) MRI.
- MRtrix - a set of tools to perform diffusion-weighted MR white-matter tractography in a manner robust to crossing fibres, using constrained spherical deconvolution (CSD) and probabilistic streamlines.
- Virtual MRI - a realistic simulation of an MRI scanner.
Reference
2- Bio-Linux
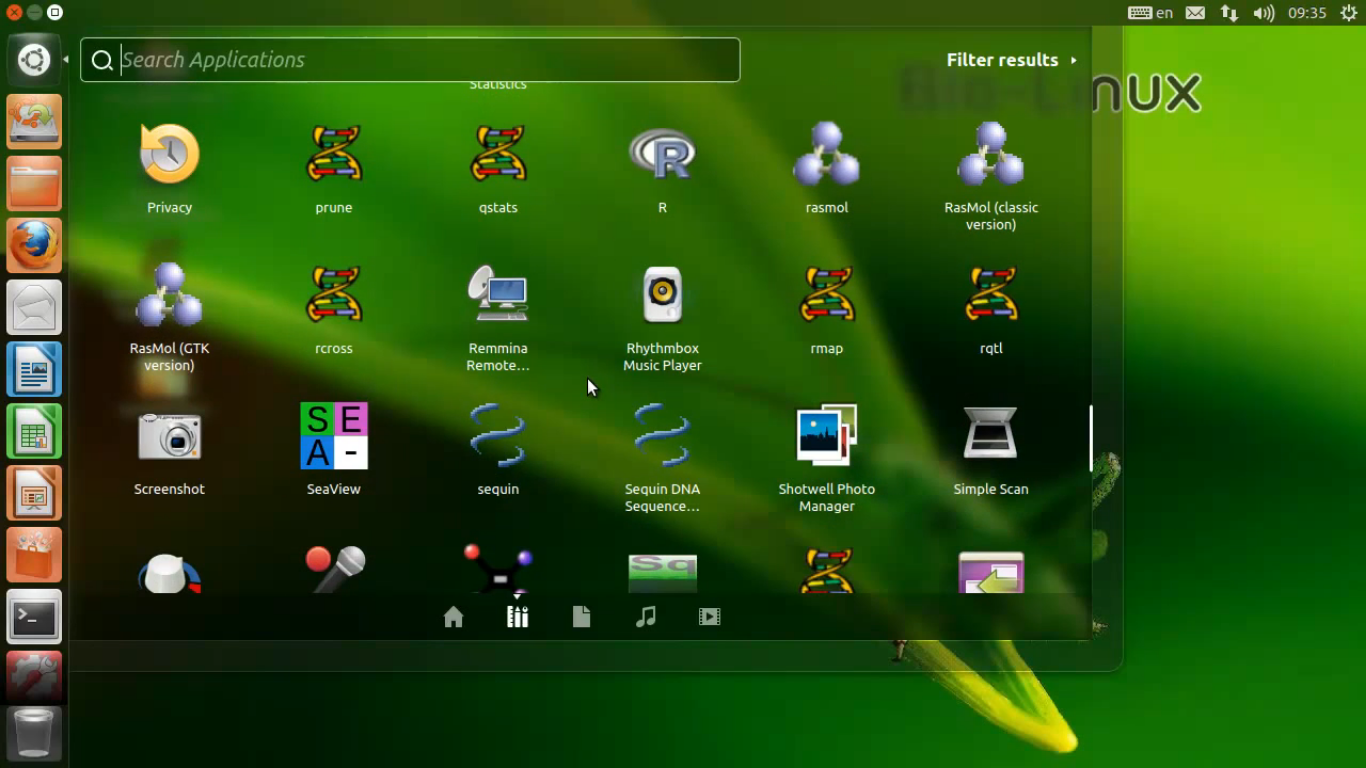
Bio-Linux is a powerful, free bioinformatics workstation platform that can be installed on anything from a laptop to a large server, or run as a virtual machine. Bio-Linux adds more than 250 bioinformatics packages to an Ubuntu Linux base, providing around 50 graphical applications and several hundred command line tools. The Galaxy environment for browser-based data analysis and workflow construction is also incorporated in Bio-Linux.
Applications
There are more than 250 bioinformatics packages, some examples are:
- abyss - de novo, parallel, sequence assembler for short reads.
- archaeopteryx - a phylogenetic tree viewer and editor.
- artemis - free genome viewer and annotation tool.
- axiome - QIIME and mothur automation toolkit.
- bamtools - toolkit for manipulating BAM (genome alignment) files.
- big-blast - The big-blast script for annotation of long sequences.
- blast2 - Basic Local Alignment Search Tool.
- bowtie2 - ultrafast memory-efficient short read aligner.
- catchall - Analyze data about microbial species abundance.
- emboss - european molecular biology open software suite.
- estscan2 - detects coding regions of DNA sequences.
- fasta - Collection of programs for searching DNA and protein databases.
- galaxy-server - Web-based analysis environment for bioinformatics.
- jalview - multiple alignment editor.
- lucy - Preparation of raw DNA sequence fragments for sequence assembly.
- mothur - sequence analysis suite for research on microbiota.
- ncbi-blast+ - next generation suite of BLAST sequence search tools.
- rasmol - Visualize biological macromolecules.
- ssake - genomics application for assembling millions of very short DNA sequences.
- swarm - robust and fast clustering method for amplicon-based studies.
Reference
3- Fedora Robotics
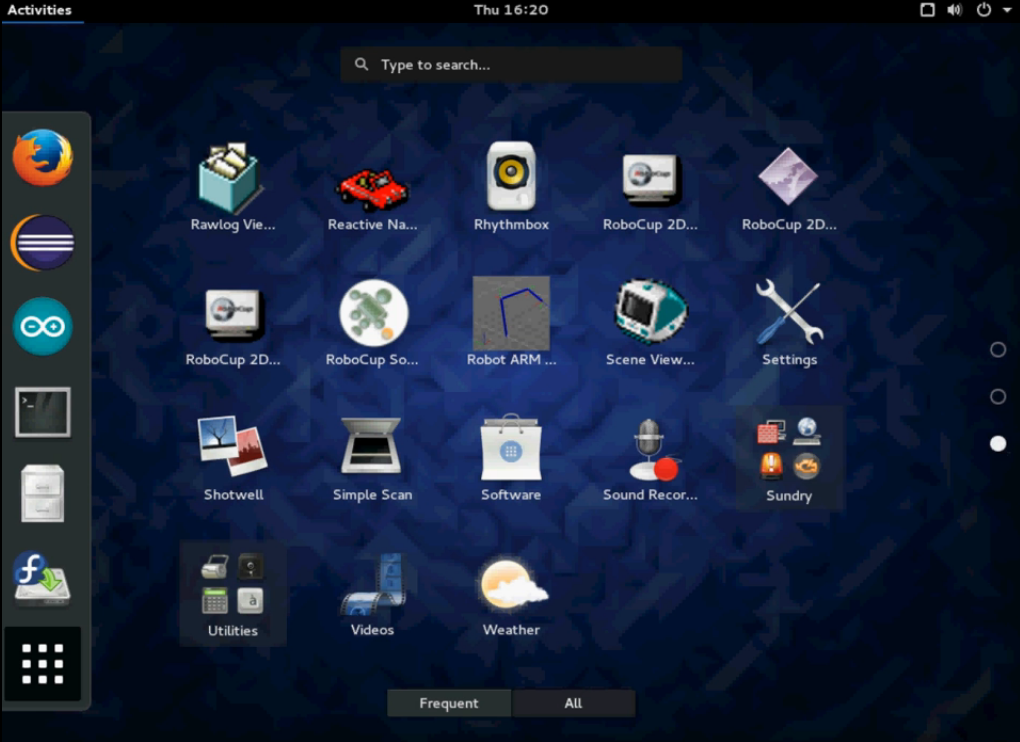
Fedora Robotics spin provides a wide variety of free and open robotics software packages. These range from hardware accessory libraries for the Hokuyo laser scanners or Katana robotic arm to software frameworks like Fawkes or Player and simulation environments such as Stage and RoboCup Soccer Simulation Server 2D/3D. It also provides a ready to use development environment for robotics including useful libraries such as OpenCV computer vision library, Festival text to speech system and MRPT.
Applications
- Player - Robot server and framework for advanced robotic systems.
- Gazebo - A versatile and powerful 3D robotics simulator with realistic physics.
- Arduino - Development environment for the Arduino open source hardware platform.
- Fawkes - Modular framework for high performance robotic systems.
- SimSpark - Physical simulator for multi-agent robotics research.
- Stage - A flexible 2.5D simulator for robotic sensors and movement.
- PCL - a standalone, large scale, open project for 2D/3D image and point cloud processing.
- MRPT - Libraries and applications for robotics system development.
- Eclipse - Powerful and extensible multi-language IDE.
Reference
4- Fedora Astronomy
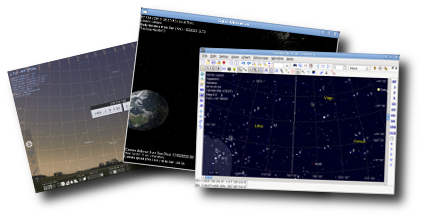
Fedora Astronomy provides the Fedora KDE desktop enhanced with a complete scientific Python environment and the AstrOmatic software for data analysis. KStars was added to provide a full featured astrophotography tool. As KStars uses the INDI library to control equipment, various telescopes, cameras etc. are supported.
Applications
- Astropy - a community python library for Astronomy to foster interoperability between astronomy packages.
- Virtualplanet - allows you to visualize any planet aspect at any time.
- Kstars - provides an accurate graphical simulation of the night sky, from any location on Earth, at any date and time.
- Siril - a free astronomical image processing software.
- Celestia - The free space simulation that lets you explore our universe in three dimensions.
- INDI - a cross-platform software designed for automation and control of astronomical instruments.
- AstrOmatic - a set of astronomical pipeline software.
- GIMP - a powerful image composition and graphic editing app, similar to Adobe® Photoshop™.
Reference
5- Poseidon Linux

Poseidon Linux is based on Ubuntu (previously in Kurumin/Knoppix) and aimed at the international scientific community (students, technicians, professors, researchers). It offers several specific tools in the areas of Bathymetry, Seafloor Mapping, GIS and 3D Visualization. The main software are:
- MB-System - an open source software package for the processing and display of bathymetry and backscatter imagery data derived from multibeam, interferometry, and sidescan sonars.
- GMT (Generic Mapping Tools) - an open source collection of about 80 command-line tools for manipulating geographic and Cartesian data sets.
- GIS packages.
Reference
6- Debian Med

Debian Med is not a stand-alone linux distro, but a collection of packages for medical practice and biomedical research, that can be installed in your Debian or Debian-derivative system. It contains 1039 packages which are grouped by metapackages. Each metapackage will cause the installation of packages for a specific topic. Debian Med is working closely together with Bio-Linux.
Applications
- Biology - bioinformatics packages.
- Biology Development - packages for development of bioinformatics applications.
- Next Generation Sequencing - bioinformatics applications usable in Next Generation Sequencing.
- Phylogeny - phylogeny packages.
- Cloud - bioinformatics applications usable in cloud computing.
- Content Management - content management systems.
- Medical data - drug databases.
- Dental - packages related to dental practice.
- Epidemiology - epidemiology related packages.
- Hospital Information Systems - Debian Med suggestions for Hospital Information Systems.
- Imaging - image processing and visualization packages.
- Imaging Development - image processing and visualization packages development.
- Laboratory - Debian Med suggestions for medical laboratories.
- Oncology - packages for oncology.
- Pharmacology - packages for pharmaceutical research.
- Physics - packages for medical physicists.
- Practice - packages for practice management.
- Psychology - packages for psychology.
- Rehabilitation - packages for rehabilitation technologies.
- Research - packages for medical research.
- Statistics - packages for statistics.
- Tools - Debian Med several tools.
- Typesetting - support for typesetting and publishing.
Reference
7- NeuroDebian

NeuroDebian is not a stand-alone linux distro, but a collection of neuroscience research software that is available for Windows, macOS, Debian and Ubuntu Linux. It has a large collection of Packages for:
- Distributed Computing.
- Electrophysiology.
- Magnetic Reasonance Imaging.
- Modeling of neural systems.
- Neuroscience Datasets.
- Neuroscience Education.
- Psychophysics.
Reference
Conclusion
The aforementioned choices cover diverse specialties, and there are also other linux distros targeting medical and scientific communities but are not actively developed anymore, like Scientific Linux, OSDDlinux and BioSLAX. Linux world is vast and everyone can find the distro that suits his/her specific needs or even customize it if he/she has the knowledge to do so.
Art source


