BrainBrowser: Open source Web-based Brain Visualization with Volume & Surface viewers
BrainBrowser is an open source JavaScript Library project aiming to provide 3D brain visualization through the web browser. It uses web technologies like HTML5, WebGL and web-ready 3D visualization frameworks.
BrainBrowser is a pure Javascript project that runs directly through the browser without the need to install or configure other software like Java, Flash, or browser extensions. It depends only on browser enabled-WebGL.
Brain Browser allows The developer can display complex dataset work with few lines of JavaScript code, Without the need to expose to the basic frameworks, and technologies used like Three.JS/ and WebGL.
Highlights
- Open source, released under GPLv3
- Extensible
- Developer-friendly
- Lightweight
- Embeddable in any web projects
- Using modern technologies
- Fast even with slow networks
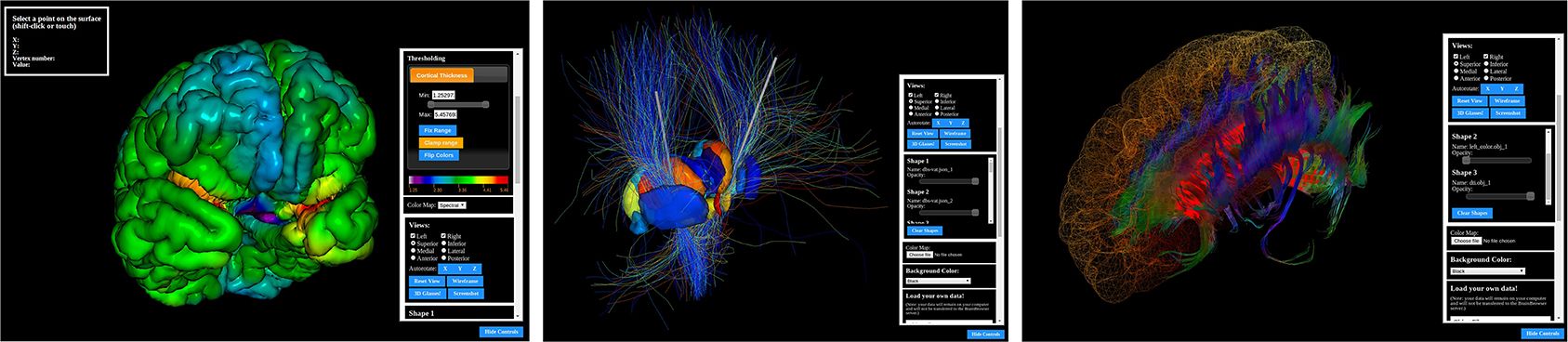
Gallery
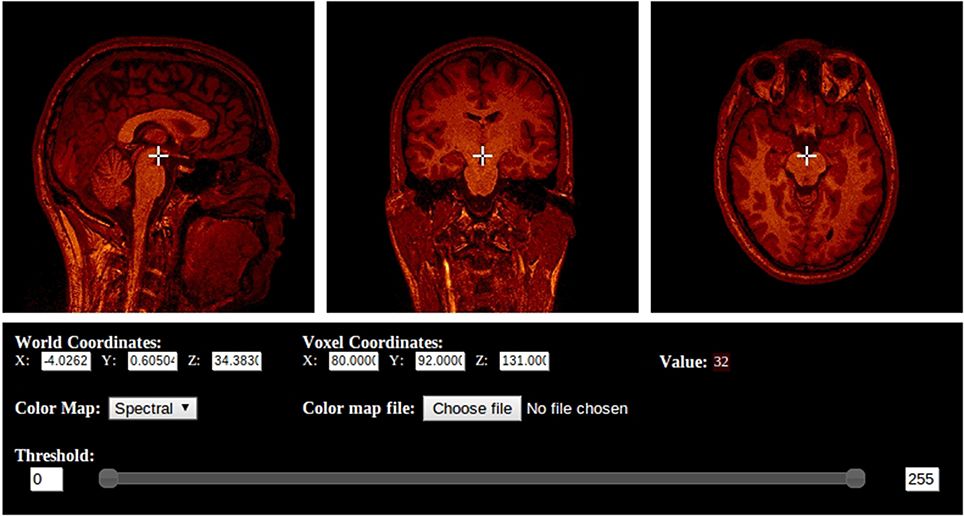
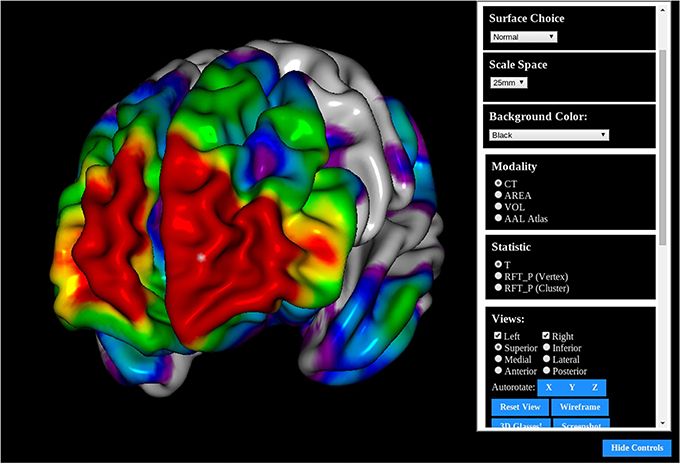
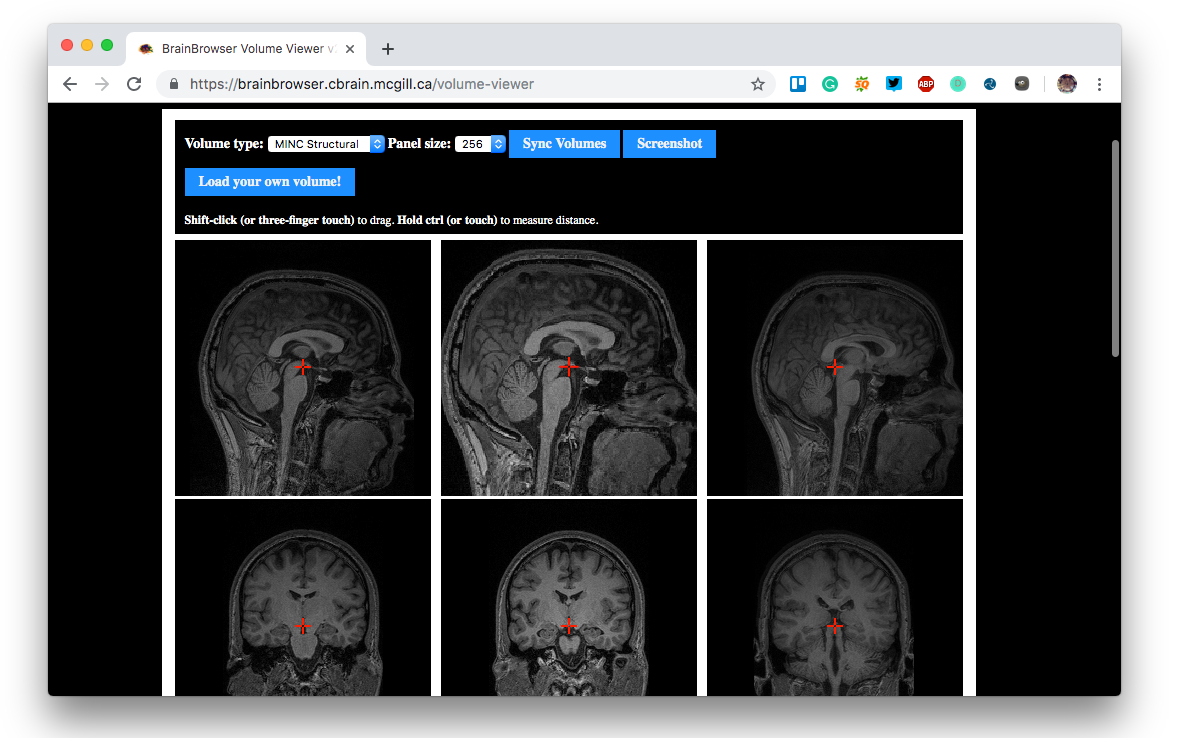
BrainBrowser is developed and maintained as part of the CBRAIN project at the McGill Centre for Integrative Neuroscience. BrainBrowser was originally created by Nicolas Kassis (and Tarek Sherif, then developed and maintained by Robert D. Vincent and Natacha Beck.
Features
- Easy to install
- Load data from networks or from local files
- Developer-ready documentation
- Developer-friendly API
- Real-time manipulation and analysis of 3D imaging
- Compatible with modern browsers (Google Chrome, Mozilla Firefox, Opera, Safari)
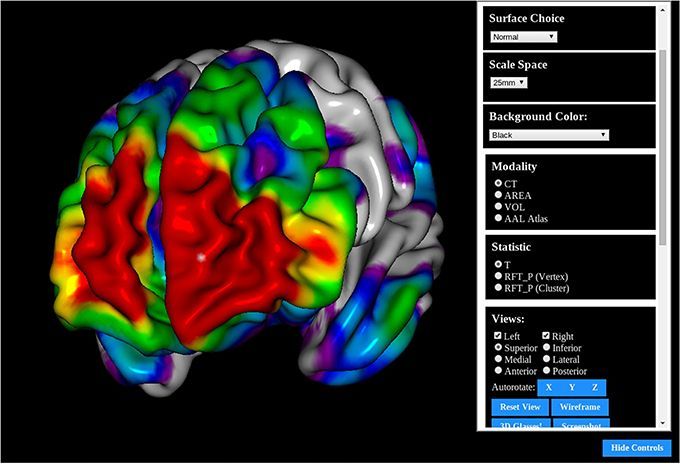
- Surface Viewer/ Surface Renderer
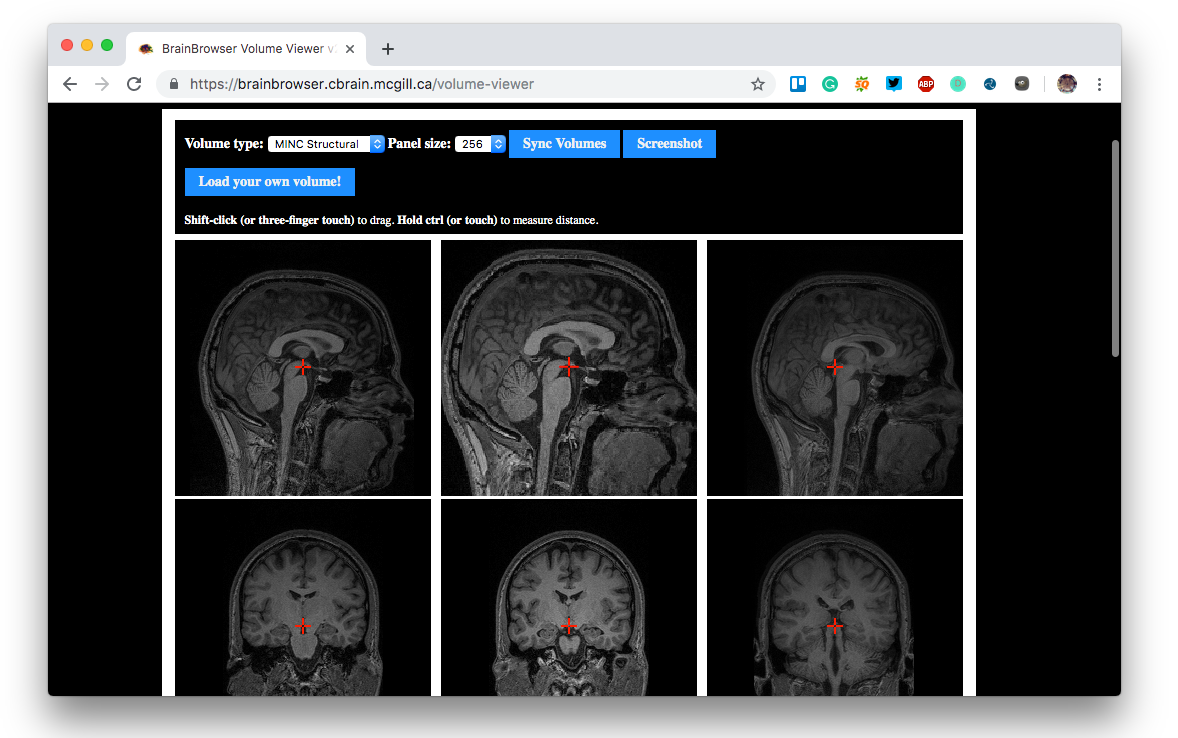
- Volume Viewer/ Volume Renderer
- MRI volume data renderer
- Navigate 3D or 4D MINC structural or functional MRI volume data
- Volume Viewer that simultaneously displays slices along the saggital, tranverse and coronal planes
- MACACC Dataset-ready
- Deep brain stimulation data exported from LEAD-DBS in JSON format
- Cortical amyloid deposition of a cohort of 18 month-old transgenic mice with Alzheimer's disease
- 3D structural data (MINC MRI data)
- 4D functional data (MINC MRI data)
- Web-services support
Technologies
- NodeJS
- Three.JS: a cross-browser JavaScript library and Application for building 3D browser-ready objects/ animation with support of WebGL.
Web-Service
Web-service is an API that uses local data to render 3D brain model within an embedded 3D viewer into any webpage with one request call. It's fairly customizable through a callback function.
https://brainbrowser.cbrain.mcgill.ca/surface-viewer-widget
Please note the Surface Viewer Web Service is still in its early stages of development. Performance is not guaranteed and the API may change rapidly.
Resources
- BrainBrowser's page
- BrainBrowser [Github]
- BrainBrowser [Demo]
- BrainBrowser [Poster - PDF]
- [Paper] BrainBrowser: distributed, web-based neurological data visualization
- Tarek Sherif (Developer)
- Nicolas Kassis (Developer)