CamiTK, A FREE Open Source Framework to easily and rapidly prototype CAMI Applications
CamiTK (Computer Assisted Medical Intervention Tool Kit) is a Free Open Source Application and Framework developed at the TIMC-IMAG laboratory of the Grenoble University, France. It helps researchers and clinicians to easily and rapidly collaborate in order to prototype CAMI applications, that feature medical images, surgical navigation and biomechanical simulations.
It was developed internally since 2001 and the first open source version was version 2.0 which released in November 2011. It is developed with C++, Qt and VTK.
CamiTK Framework
It consists of three components: CamiTK Core, CamiTK Service Layer and CamiTK Extensions.
1- CamiTK Core
- Definition and Implementation of concepts.
- Only experts are supposed to understand it well.
2- CamiTK Service Layer
- The glue between your problem and the framework.
- Developers need basic knowledge.
3- CamiTK Extensions
- Where domain-specific work is done.
- Focus on your project/problem.
Knowledge gathering
CamiTK avoids reinventing the wheel by gathering the knowledge and know-how from several research fields:
1- Perception
- Visualization
- Interaction
- Processing
- Analysis
2- Reasoning
- 3D geometries
- Interaction
- Biomechanics
3- Action
- Tracking
- Navigation
- Robot Control
Some projects based on CamiTK
1- ROBACUS
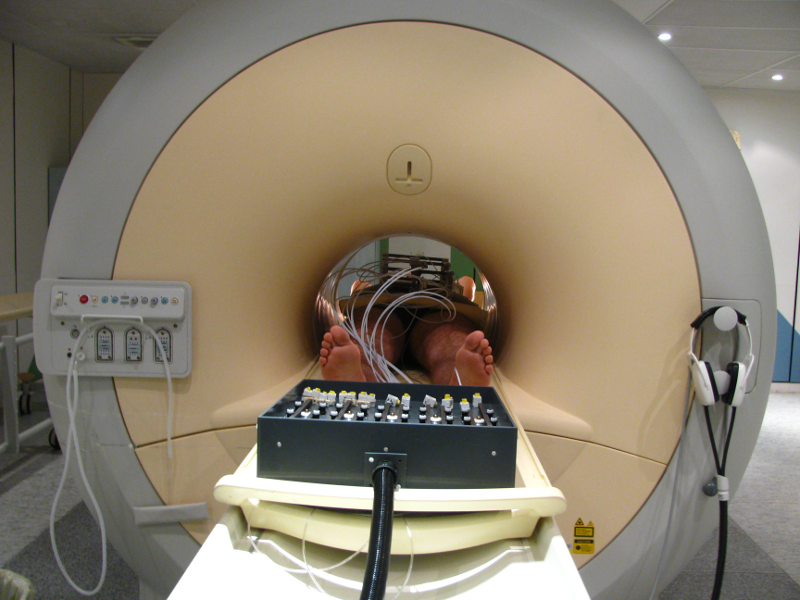
ROBACUS is a patient-mounted CT and MRI guided interventional radiology robot for percutaneous needle interventions. The 5 degree of freedom robot uses ultrasonic motors and pneumatics to position the needle and then insert it progressively. The needle position and inclination can be registered in the images using two strategically placed fiducials visible in both imaging modalities. The robot is controlled through CamiTK as a component with a set of actions that can be applied to make it move. The image processing for fiducial segmentation and registration is done using existing CamiTK components.
2- Instument tracking via condensation algorithm - Urology
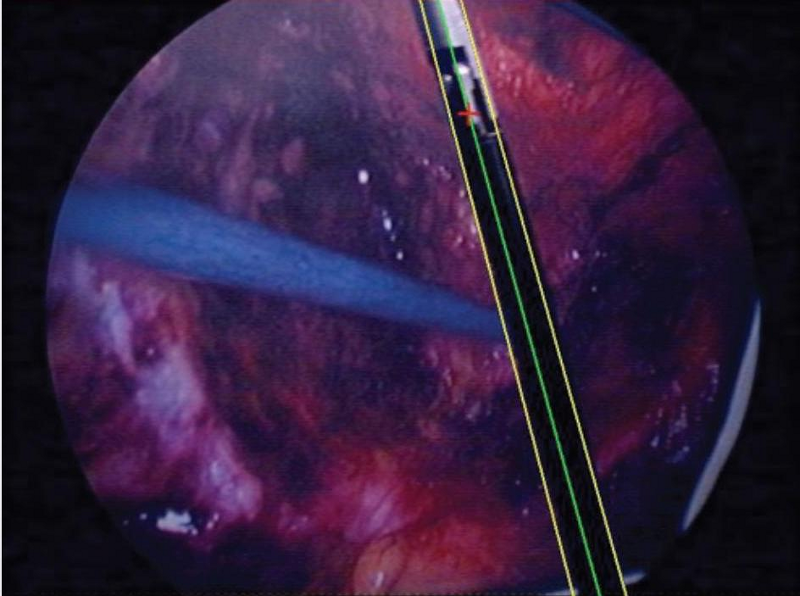
This project allows instrument tracking using 2D laparoscopic images. The instrument's orientation is detected using the CONDENSATION algorithm (M. Isard et A. Black, Conditional Density Propagation for Visual Tracking). General steps can be re-implemented using CamiTK (particle choice, particle evolution, ...). We can then create our own framework by defineing our own instrument with its own characteristics and then track its 3D orientation using the known insertion point.
3- Light Aspiration device for in vivo Soft Tissue Characterization (LASTIC)
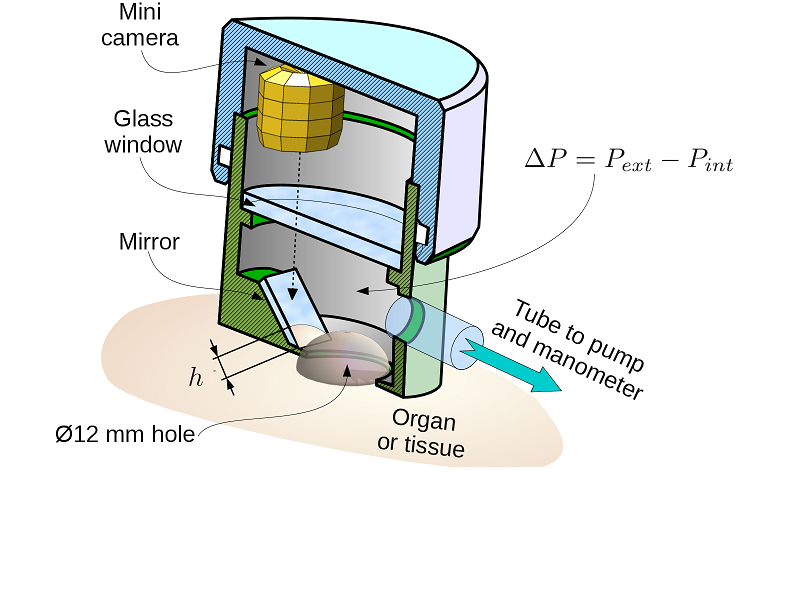
LASTIC is a measurement tool used to determine soft tissue elasticity, a key parameter for the simulation and the deformation of organs and soft tissues (like the tongue or the skin). This device consists of three components: a syringe-pump to generate a depression, a manometer to measure the depression and a camera inside LASTIC to visualize the deformation of the studied soft tissue. This project integrates a CAMITK library to communicate through serial ports to control the manometer and syringe-pump.
4- MML framework
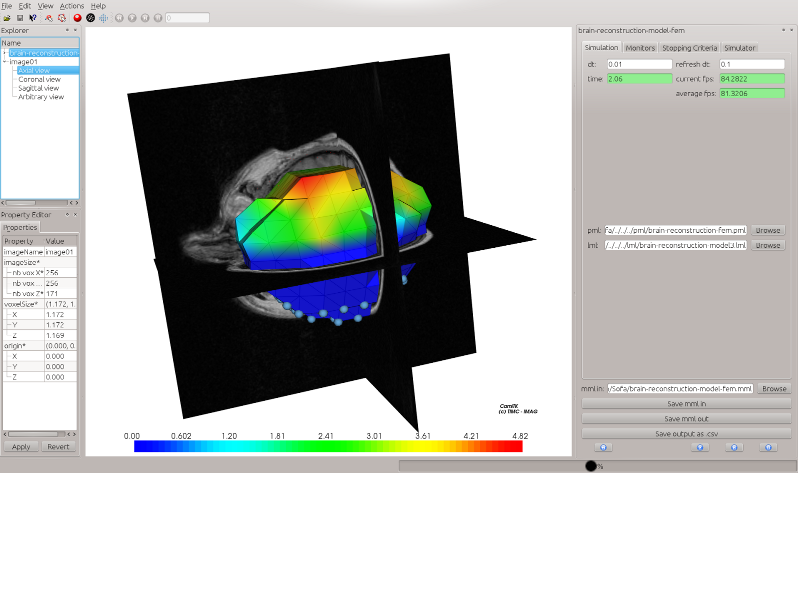
The MML framework is designed to help scientists in the difficult problem of evaluating and comparing bio-mechanical models of various types (e.g. FEM, Mass-spring) and from different simulation engines (e.g. Sofa, Artisynth, Ansys).
5- Biomechanical Modeling
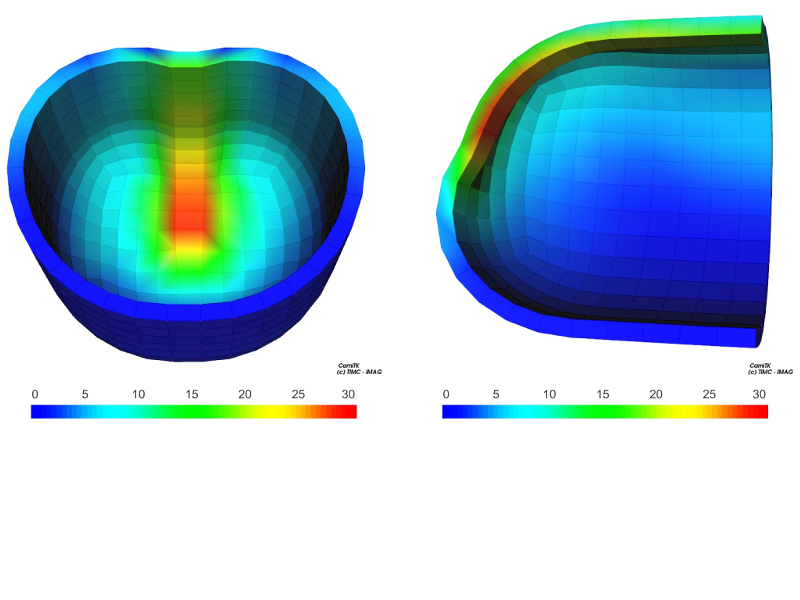
CamiTK can be used to create 3D models comprised of multiple deformable objects. Various parameters (elasticity, contraction, incompressibility...) can be implemented on the deformable objects, in order to monitor how they affect the behavior of certain particles/objects/entire structure. The tool to vizualize the model in a 3D environment is camitk-imp. A program to convert an xml document executable by CamiTK to an xml document executable by SOFA (valid only for specific usage) has also been created. Finally, it is possible to visualize in camitk-imp a model initially created for the SOFA simulator using MML.
6- BiopSym
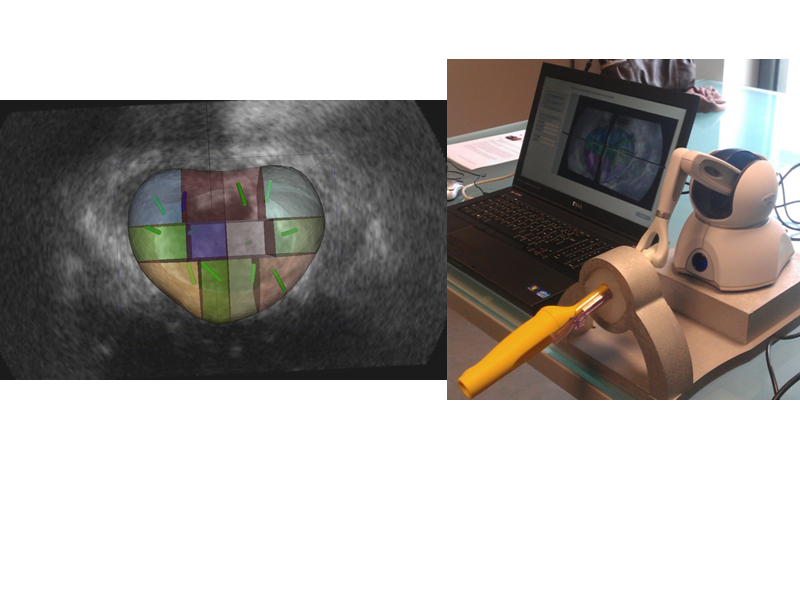
BiopSym is a virtual reality simulator with a learning environment for image-guided prostate biopsy. The simulator,a laptop computer connected to a haptic device with a stylus held as the ultrasound probe, includes a clinical case database (three dimensional ultrasound volumes) and a learning environment with specific exercises and a module to replicate a randomized 12-core biopsy procedure. A visual (biopsy mapping) and numerical feedback (score) is given to the user.
Highlights
- Free and open source.
- Cross platform.
- Provides fast and stable tools for prototyping medical applications.
- Fast technological transfer between students, PhD, research scientists, clinicians and industrial partners.
- Easy integration of algorithms, data, and devices.
- Knowledge gathering.
- Support for common medical imaging and biomechanical model formats (DICOM, VTKImage, ITKImage, VTKMesh, PML, VML).
- Greatly modular with an easy extension mechanism that clearly separates domain logic (data and algorithms) from application logic (visualization and interaction).
Developers
There are three main developers: Celine Fouard, Emmanuel Promayon (they are both teacher-researchers at the TIMC-IMAG laboratory of the Grenoble University in the CAMI team) and Jean-Loup Haberbusch (joined the team as research and development engineer to work full-time on the project). There are many other contributors to the project as well.
License
- LGPL v3.0


