MatchMiner: An Open-source Computational platform for Clinical Trials Genomic Matching
Table of Content
The challenge of matching precipitant (patients) for cancer trials is not easy nor simple. The current methods for patient recruitment for clinical trials result in failure [5]. Our topic of the day MatchMiner is designed to help researchers overcome this challenge.
MatchMiner is an open-source computational platform with a specific focus on patient genomic profiles to precision cancer medicine clinical trials. It is intended for researchers with software development skills.
The project is developed by researchers at Dana-Farber Cancer Institute (DFCI) which is known as a comprehensive cancer treatment and research institution in Boston, Massachusetts, United States. It's also an affiliate of Harvard Medical School.
What is Clinical Trial Markup Language (CTML)?
The Clinical Trial Markup Language or (CTML) is a new standard that aims to make a new standard for structured clinical trial data.
It is introduced by Ethan Siegel in his article "Making Clinical Trial Eligibility Machine Readable". Siegel is also one of the core researchers who developed MatchMiner.
Here are two examples captured from Siegel's article:

MatchMiner is formed of 3 parts:
- MatchMiner engine
The engine is responsible for parsing structured clinical trial data. It follows the Clinical Trial Markup Language (CTML) Standard. - MatchMiner API
The API is built as an interface for the engine. It is built with Python3.6, uses Eve framework and MongoDB a NoSQL database engine for data storage. - MatchMiner UI
The MatchMiner UI is a front-end application that is built on top of the API. It's written in JavaScript (NodeJS 10.16.9) and uses AngularJS. It uses Gulp for building and deployment.
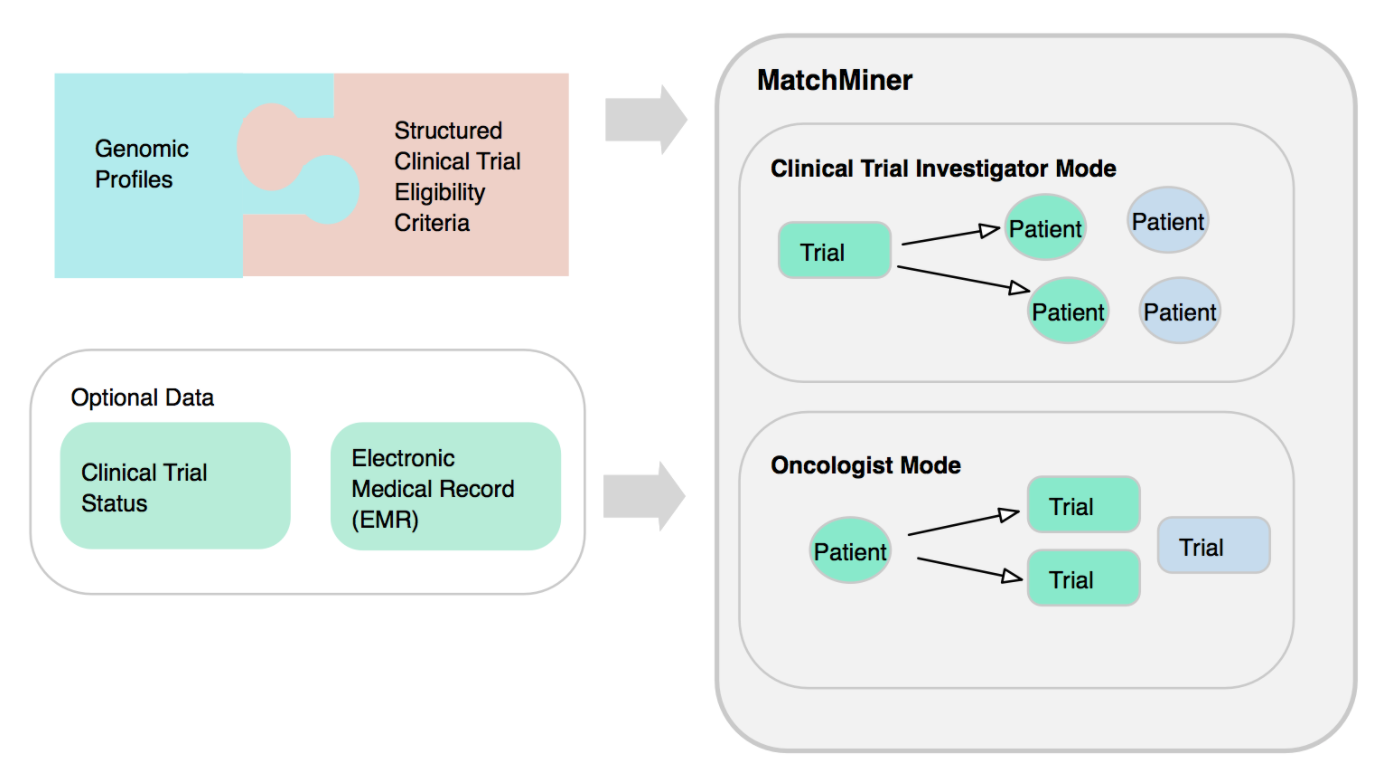
MatchMiner UI offers user authentication, user roles management, and two modes: a clinical trial investigator mode, and oncologist mode.
How does MatchMiner work?
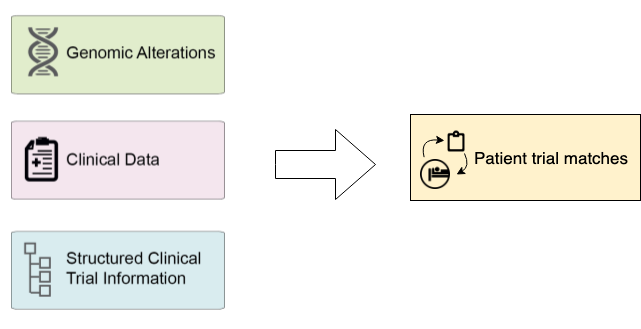
It uses patient records extracted from the electronic medical record (EMR) or electronic health record (EHR) systems and clinical trial data in Clinical Trial Markup Language standard.
The MatchMiner platform matches patient-specific genomic events to clinical trials, and makes the results available to trial investigators and clinicians via a web-based platform.
Features
- Open-source
- Complete API support
- Developer-friendly
- Extensible though plugins and extensions
- Well written documentation
- Command-line interface
- Elasticsearch support
Platforms
MatchMiner can be installed in any platform that has Python and NodeJS.
Technologies used
- Python3.6
- NodeJS
- Eve Rest Framework
- MongoDB
- Angular Framework
- Gulp
- Docker
License
MatchMiner is released as an open-source project under Apache License 2.0.

Conclusion
Development-wise, MatchMiner follows a good design pattern to ensure a stable and fixable product. Splitting the engine from the user-interface is a good decision as well as proving a headless REST-API on top of the engine which allows developers to integrate it easily in their projects, or build their own applications on top of it.
It's an approach we rarely see from developers with research background as many tend to use old-school approaches to solve their research computational problems.
Resources
- Official Website: https://matchminer.org/
- MatchMiner: Documentation
- https://github.com/dfci/matchminer
- https://www.dana-farber.org/
- https://medium.com/matchminer/making-clinical-trial-eligibility-machine-readable-3fd3dffb9a8b
- https://medium.com/matchminer/clinical-trial-matching-at-the-dana-farber-cancer-institute-f4c57ac18ab0
- https://cancerres.aacrjournals.org/content/80/16_Supplement/3382
- https://blog.dana-farber.org/insight/2017/01/how-do-i-find-the-right-clinical-trial-for-me/
- https://www.hbs.edu/news/releases/Pages/matchmaker-wins-hbs-kraft-challenge.aspx
Photo by Anna Shvets from Pexels