10 Open-source Whole-Slide Image Viewers and Analysis Programs; Redefining Digital Pathology
What is Whole-slide image or Virtual Slides?
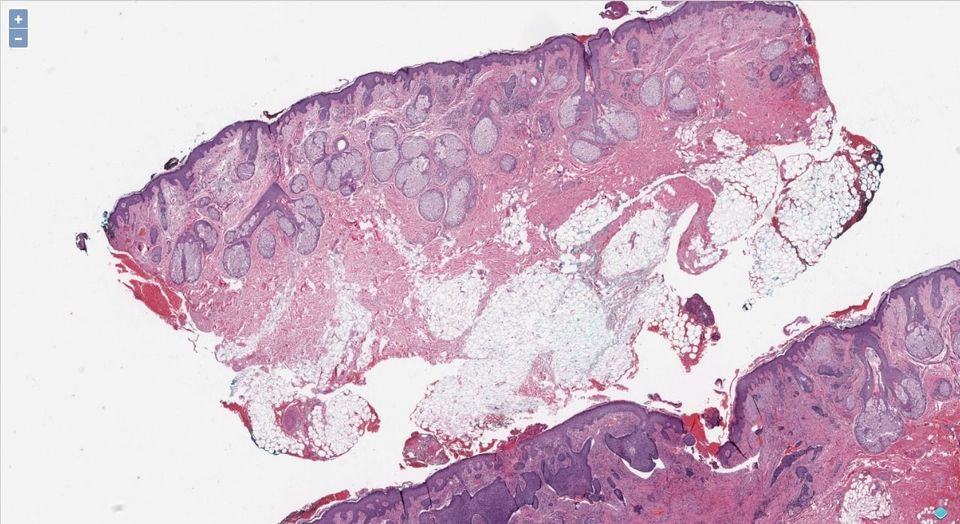
Whole-slide images (Virtual Slides), are a high-resolution image of physical histopathology scanned with special scanners, that produces the images in gigapixels. It allows scientists, researchers, clinical pathologists to use software to zoom for regions of interest ROIs.
Whole-slide images viewers are used in different sectors as medical diagnosis, intraoperative diagnosis, medical education, medical students training, diagnostic decision support systems, clinical research, & immunohistochemistry (IHC).
Whole-slide image and digital pathology
We often hear the term digital pathology, which is a technical concept to improve the quality, reduce the errors, and provide a better view of the slides. Above all Digital pathology is aiming to provide collaborative methods to improve diagnosis and research.
Whole-Slide Image formats
There are several Whole-slide image formats, there is no standard version because the variety of vendors, but we have libraries, projects, software which are built to process those formats, here is a list of the most common formats [src: OpenSlide]:
- Philips (.tiff): single-file pyramidal tiled TIFF or BigTIFF with non-standard metadata
- Aperio (.svs, .tif): single-file pyramidal tiled TIFF, with non-standard metadata and compression
- Hamamatsu (.vms, .vmu, .ndpi): multi-file JPEG/NGR with proprietary metadata and index file formats, and single-file TIFF-like format with proprietary metadata
- Leica (.scn): single-file pyramidal tiled BigTIFF with non-standard metadata
- Sakura (.svslide): SQLite database containing pyramid tiles and metadata
- MIRAX (.mrxs): multi-file with very complicated proprietary metadata and indexes
- Trestle (.tif): single-file pyramidal tiled TIFF, with non-standard metadata and overlaps; additional files contain more metadata and detailed overlap info
- Ventana (.bif, .tif) single-file pyramidal tiled BigTIFF with non-standard metadata and overlaps
- Generic tiled TIFF (.tif): single-file pyramidal tiled TIFF
About this list
We have compiled this list here to help researchers, students, and developers, by easing their research of finding the open-source, free whole-slide image viewers, and analysis. so, they can use freely in their work, read the code, build on it or integrate it into their current systems.
Our list includes programming libraries, software, & projects on GitHub.
Whole-slide image viewers
1- QuPath
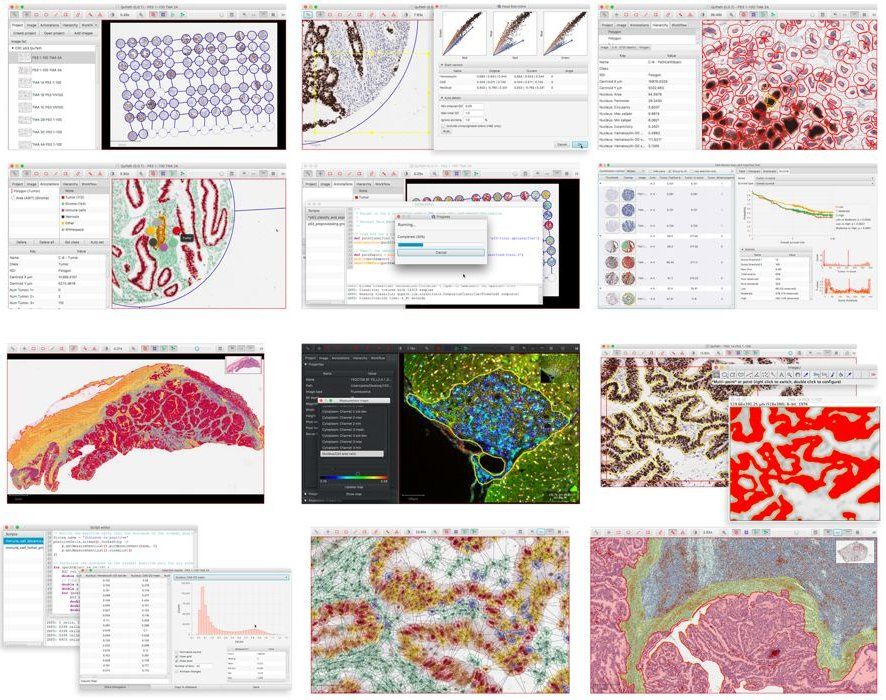
QuPath is a lightweight virtual slide viewer & analysis software that works for Windows, Linux, & macOS. It's an open-source project built using Java technologies. QuPath is developed at the University of Edinburgh.
We have provided a snap review about QuPath listing its features in this article.
2- Cytomine
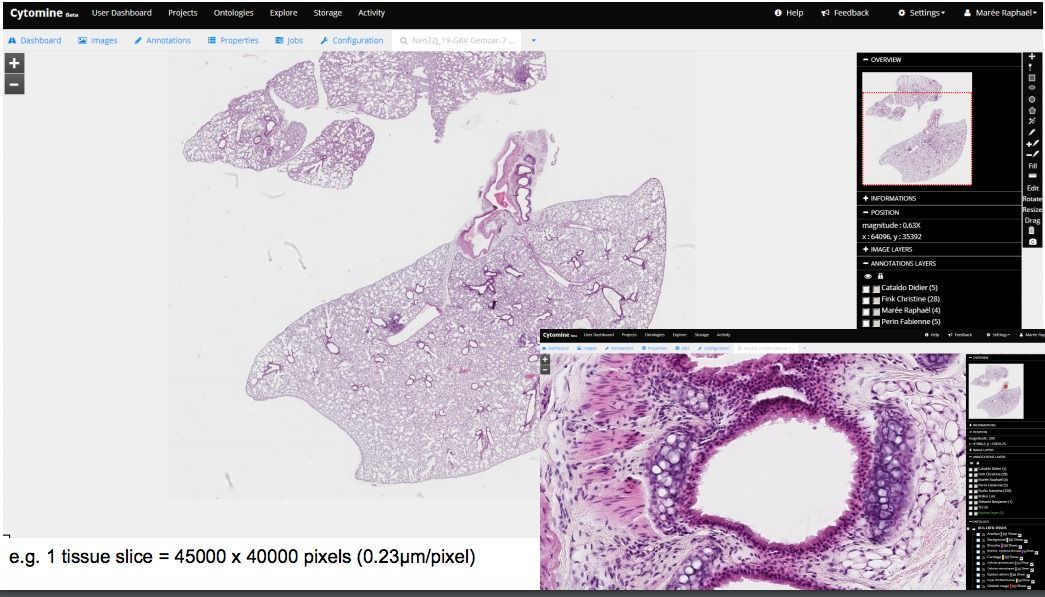
Cytomine is an open-source integrated whole-slide viewer & analysis platform. It supports many virtual slides (WSI) formats, works on Windows, Linux, macOS. With its web-based client, it supports collaboration and remote analysis for external teams, & researchers.
For more information, we have written a detailed review about Cytomine you may need to check it out.
3- Orbit
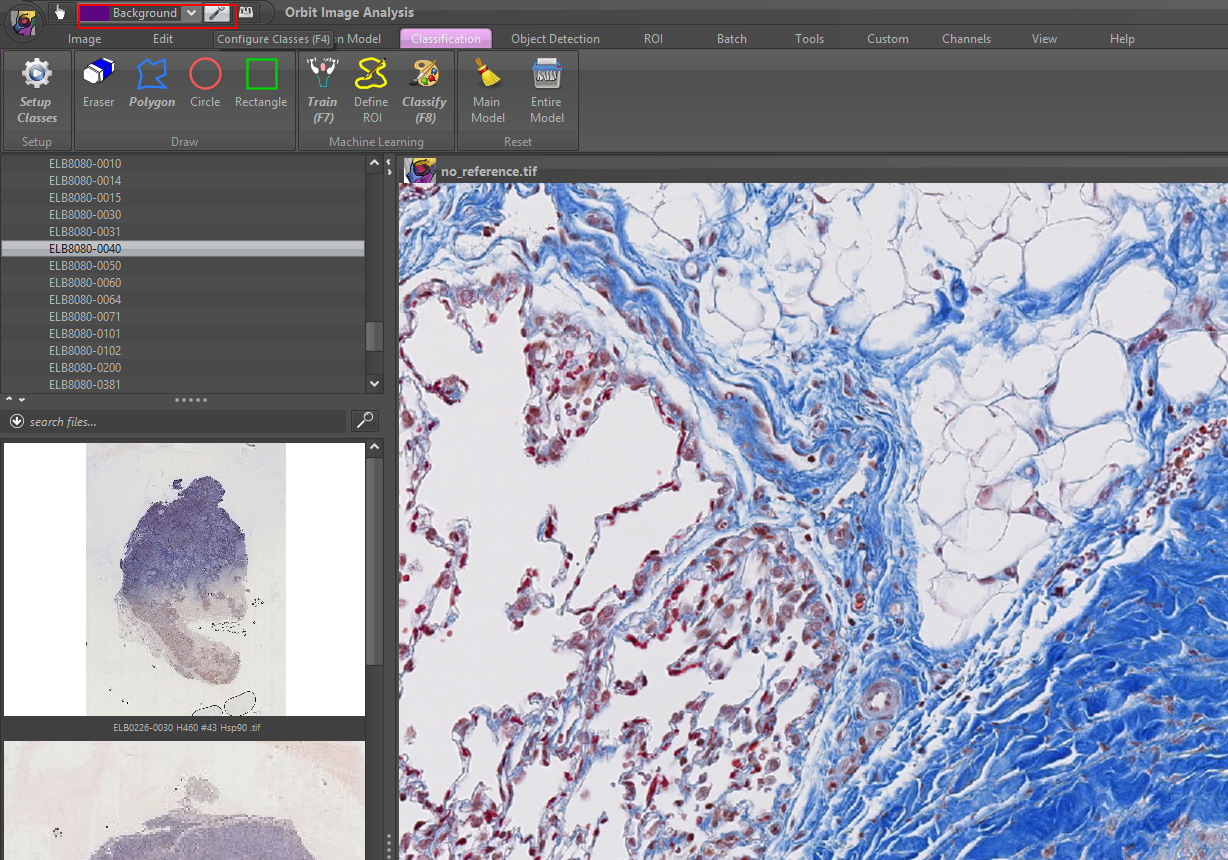
Orbit is a whole-slide image analysis program that has integrated machine, & deep learning algorithms. Orbit is built for AI researchers, but it works as a viewer for clinical pathologists as well, however, we would recommend other software for pathologists and histopathology educational purposes.
Orbit supports OMERO server, which is a digital pathology server to store, display, manage whole-slide images.
Read more about Orbit features that we covered in this article.
4- ASAP
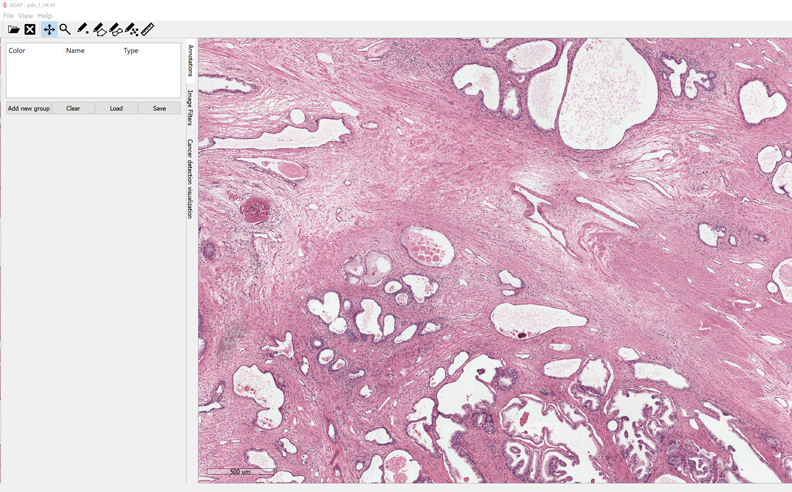
ASAP is an open-source whole-slide image viewer, it uses OpenSlide library which already supports multiple virtual slide extensions. It uses OpenCV library, DCMTK, & Boost C++ library.
ASAP is currently supporting windows (64), and Linux (Debian, Ubuntu)
5- OpenSlide with OpenSeadragon
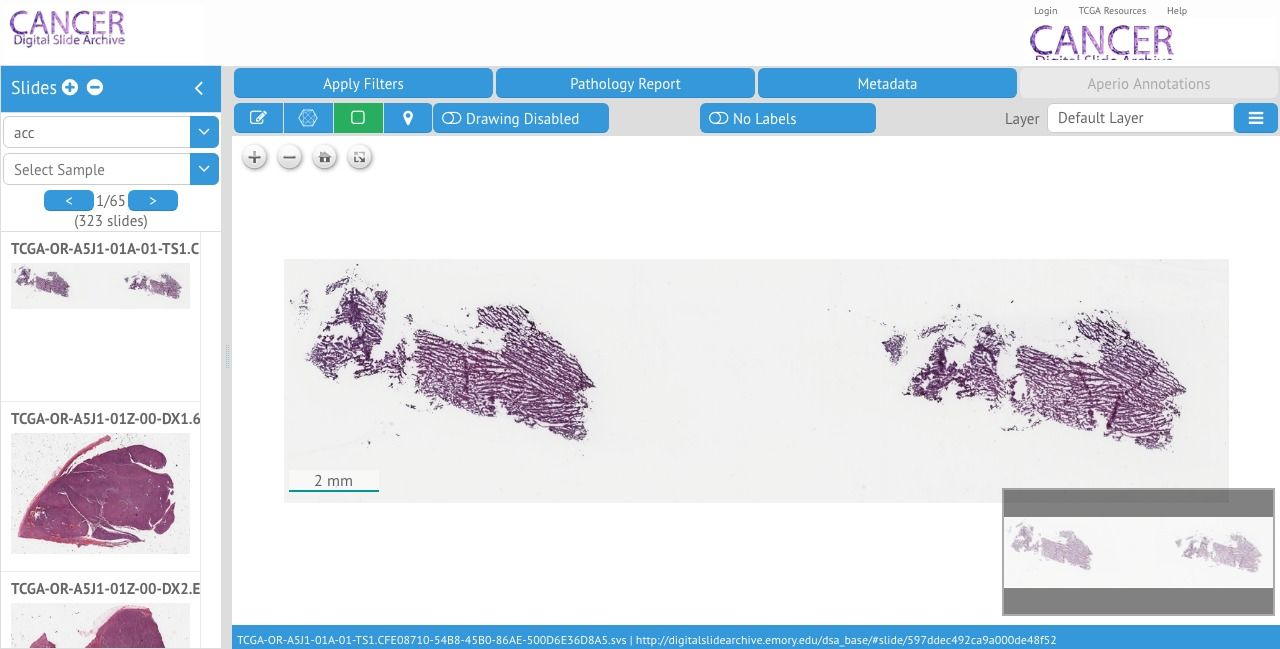
OpenSlide is an open-source C library that allows the developer to process whole-slide images (virtual slides). OpenSeadragon is a JavaScript library that allows developers to build an image viewer for high-resolution images with advanced zooming support.
Both projects have been used in several online histopathologic online projects we covered some of them while reviewing them:
6- ImageJ of SlideJs
ImageJ is a famous open-source medical imaging viewer, with a custom plugin called SlideJ, ImageJ can display whole-slide images easy, including Aperio .SVS, Leica .SCN, Hamamatsu .NDPI, Mirax, Generic tiled TIFF and regular TIFF.
SlideJ splits the large image into square tiles with user-configured settings. SlideJ is based on Bio-Formats of OME (Open Microscopy Environment)
7- OMERO viewer
OMERO is a digital pathology server designed to store, manage, whole-slide images and provide developers with API tools to build custom apps on top of it. OMERO is a production of OME (Open Microscopy Environment).
OMERO viewer is a custom whole-slide image viewer build to display images from OMERO server. It supports multiple image viewing, ROIs, annotations, and comes with advanced viewing options.
We recommend you to read our review about OME (Open Microscopy Environment) and their amazing work here.
Here is a 10 mins video that shows it in action.
8- JVSView
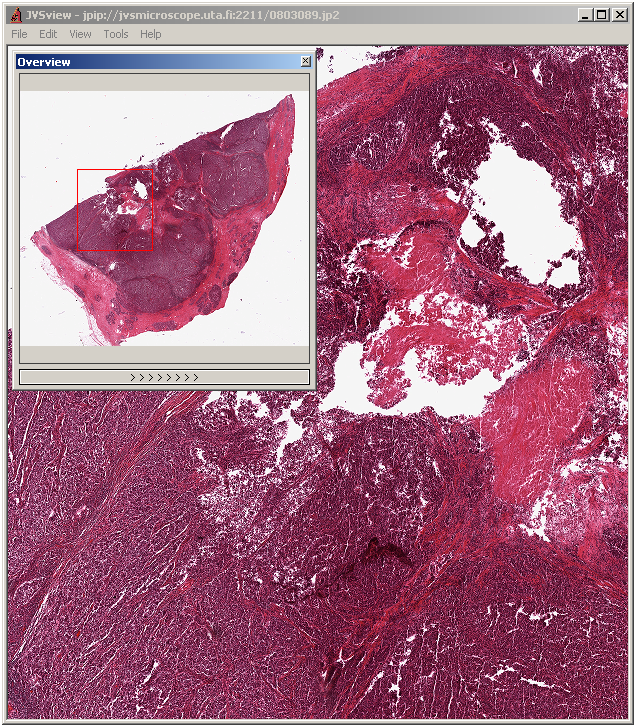
JVSView is a windows package for displaying virtual slides, it's written in C++ and works on Windows 64bits/ 32bits.
JVSView supports local and remote images from JVSserv – a JPIP server for a remote serving of JPEG2000 virtual slides. It also supports multiple full-screen displays, multi-layer viewing, storing ROIs as XML metadata. It provides seamless integration with ImageJ. It's free for non-commercial users.
9- PMA.start
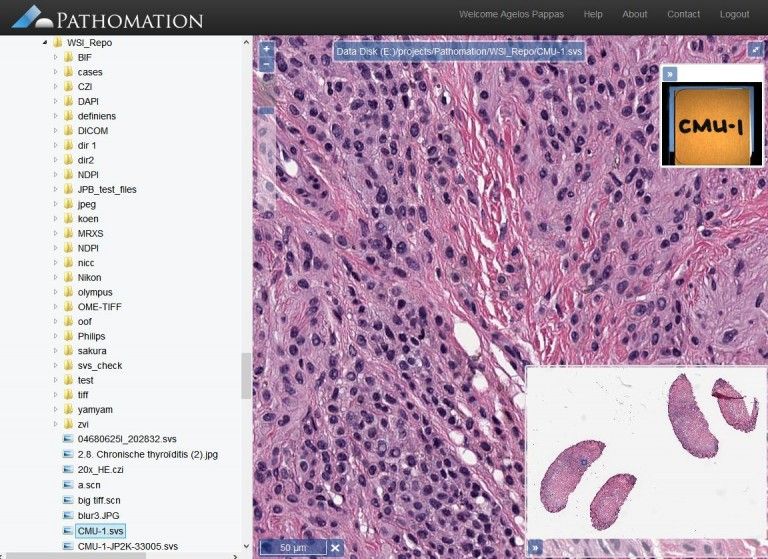
PMA.start (Pathomation) is a free whole-slide image viewer and analysis package, that comes with a modular system and extensions to support ImageJ integration, and web-services API.
Unlike several microscopy packages, Pathomation is using OpenLayers, not OpenSlide and several web technologies like jQuery, jQueryUI, Bootstrap. and it is only available for Windows.
For more information about how PMA.start works, we would recommend watching the next presentation
10- caMicroscope
caMicroscope is an open-source web-based Digital pathology image viewer with support of human/ machine-generated annotations and markups.
caMicroscope is using OpenSeadragon the JavaScript high-resolution library, & IIPImage an open-source image server designed for web-based streamed viewing and zooming of ultra-high-resolution images. It uses a tile-based method to display high-resolution images within the web browser. It also supports multiple protocols (IIP, IIIF, Zoomify, and DeepZoom protocols).
Note that caMicroscope is under active development.
Conclusion
Open-source solutions are cost-effective for labs, researchers as it allows developers and software engineers to extend it and integrate it with their apps. We have collected 30+ projects, but we choose the most mature and usable ones, hoping this list will benefit our readers.