Cytomine: Free Open source Web-based Digital Pathology (WSI) solution with Machine learning flavor
Table of Content
Cytomine is a web-based open source solution, aiming to empower whole-slide image processing, & analysis with machine learning algorithms. It's built to ease collaboration among researchers.
Cytomine is built by a group of researchers from Montefiore Institute (University of Liège, Belgium) who are developing machine learning algorithms and big data software modules aiming to provide an open-source solution for processing very large imaging data.
Unlike Orbit which we introduced in this article, Cytomine is considered lightweight, web-based, easy to install, & does not require heavy-duty hardware requirements like an orbit. It can be installed on a web server, a laptop or a desktop.
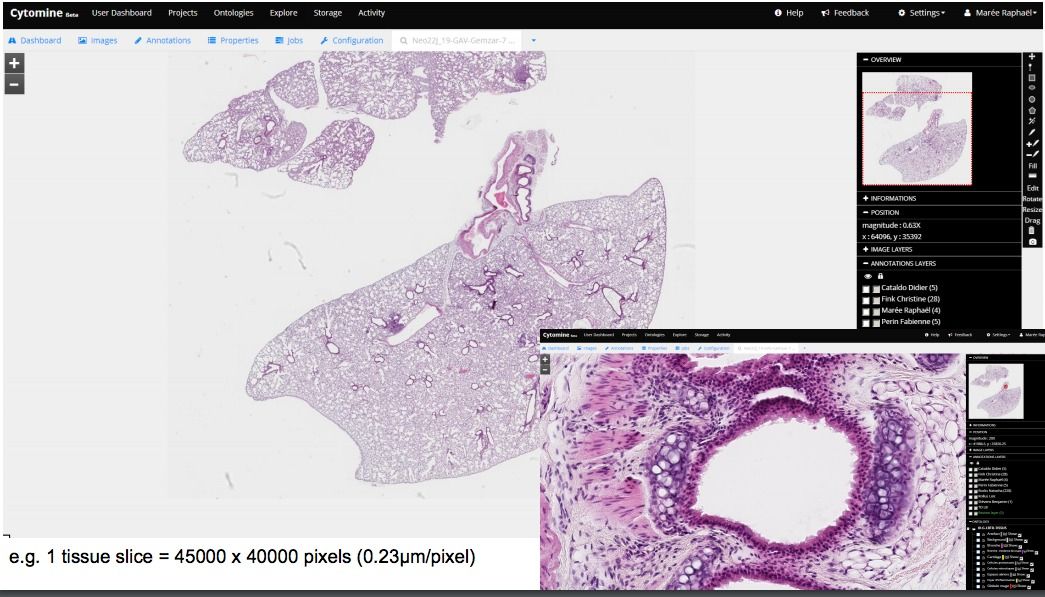
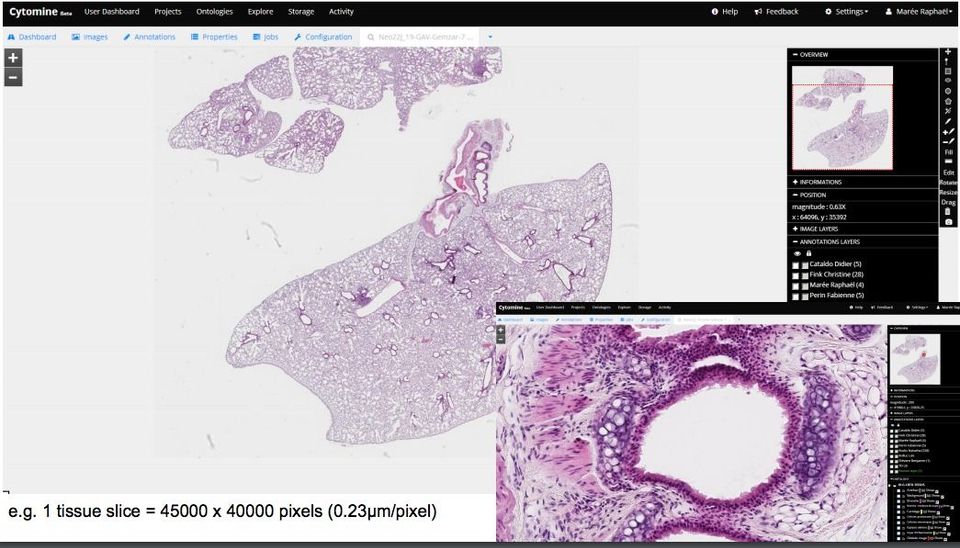
In short, Cytomin is a self-hosted cloud-based whole-slide image viewer, with multi-user, multi-projects options, empowered by machine learning algorithms, So as a web-based WSI viewer, it improves pathologists workflow, as it provides out-of-box collaboration tool, speed-up image analysis, improves the quality, and reduce the errors.
The project is meant for biologists, biomedical researchers, computer scientists, software engineers, & pathologists.
How does Cytomine work?
Before we dive into how Cytomine works, let's start by talking about the major elements in digital pathology: Whole slide scanners, and Whole slide image, but what is Digital Pathology?
Digital Pathology!
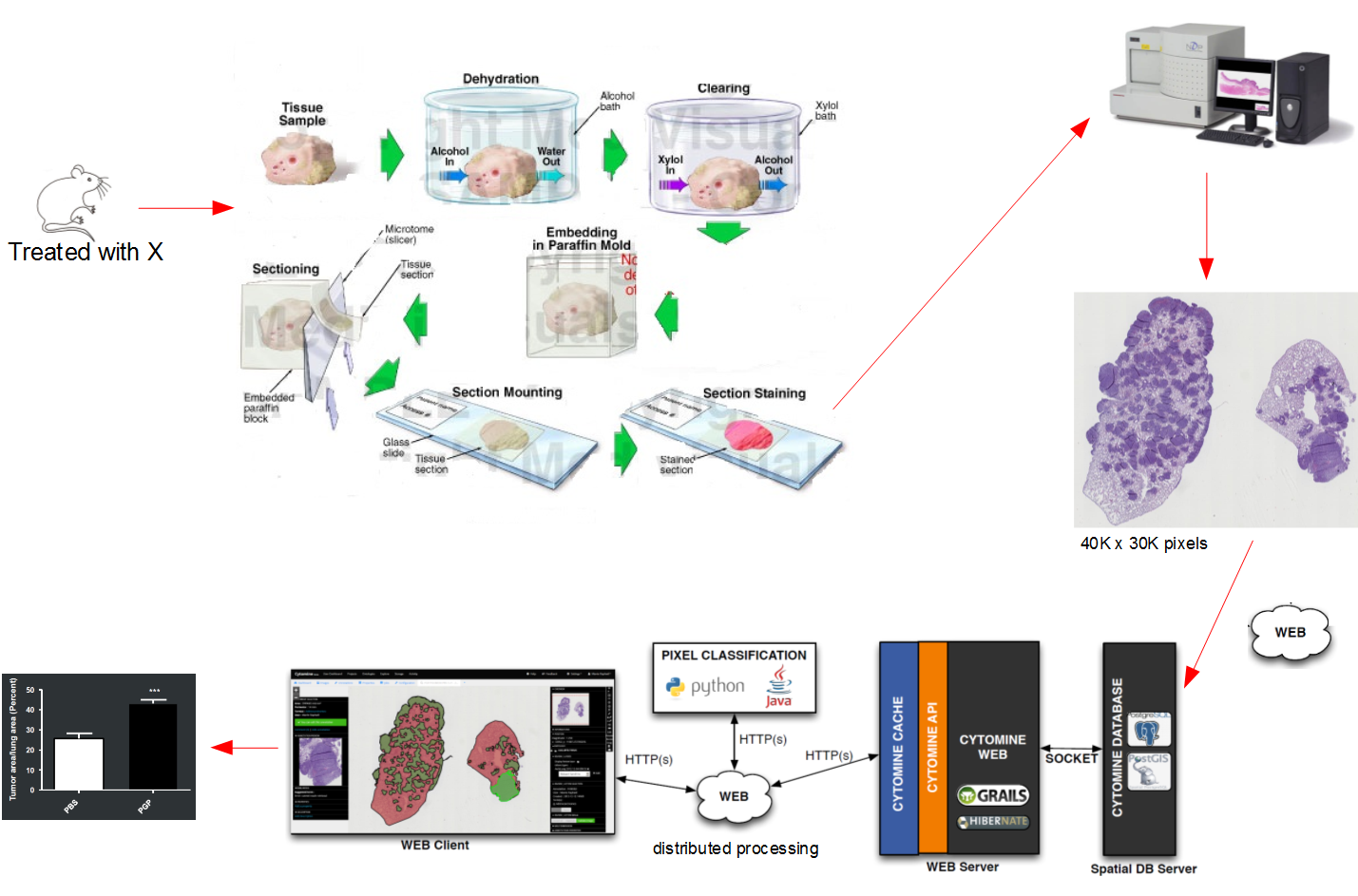
Digital Pathology is an image-based information environment that transforms the classical traditional physical glass slides or physical samples into virtual slides using Whole-image scanners. These scanners are scanning the slides to generate a detailed high-resolution large image in gigapixel.
Whole-Slide Image:
Unlike images, we capture with our phones, or cameras, Whole-slide images size is large, as used scanner (Whole-slide image scanners) that supports capturing resolutions of 0.5 microns/pixel, allowing high magnification. Opening virtual slides on the traditional image viewer programs are quite a challenge as they are not equipped large files and most of them do not support virtual slide image formats, the non-standard metadata, & the file compression used by Whole-image scanners.
- Trestle (.tif)
- JPEG 2000 (compression types 33003 or 33005)
- Leica (.scn)
- MIRAX (.mrxs)
- Sakura (.svslide) [SQL powered]
- Aperio (.svs, .tif)Ventana (.bif, .tif)
- Hamamatsu (.vms, .vmu, .ndpi)
- Philips (.tiff)
- Generic tiled TIFF (.tif)
Highlights
- Works everywhere (Java-based) (Linux, Windows, & macOS)
- Web-based
- Require developer and coding skills
- Linux/ macOS support with installation instructions
- Scriptable
- Support multiple projects
- Support multiple users/ teams
- Can be used in education, research, clinical work
Features
- Project management tools
- Remote image viewer
- Browser-based image viewer
- OpenStreetMaps browsing style (zoom in/out, pyramid tile-based)
- Storage management
- Annotation tools
- User management
- Activity manager
- Synchronized visualization of multiple modalities (given pre-registered images)
- UI visualization tool: display many images at once
- Support collaborative annotation
- ROIs (user-defined)
- ROIs supports key/value & text-based
- Generate annotation datasets
- Machine learning algorithms
- Object classification
- Live broadcasting support
- User behavior analytics modules
- Developer-friendly API (REST)
- Docker support
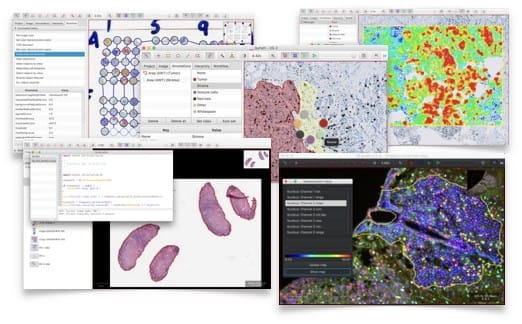
Cytomine in action
Cytomine has been used in several types of research which shows how can it be used, it can also be used for education (Histology, Pathology), you may check the research, & publication section at Cytomine website. Cytomine has ongoing researches which demonstrate it's value to open source digital pathology community, here is some of them:
- Image segmentation techniques for the quantification of whole tissue slides. (Marée et al. ISBI 2014)
- Cells sorting: detect abnormal cells for early cytological diagnosis. (Delga et al., Acta Cytologica 2014; Mormont et al., 2016.)
- Cell counting algorithms (Rubens et al.)
- Image classification and object recognition algorithms. (Marée et al., PRL 2016; ISBI 2016; Jeanray et al., PLOSOne 2015; Mormont et al. CVPRW 2018.)
- Anatomical landmark detection algorithms for morphometric change measurements e.g. in developmental and toxicological studies. (Vandaele et al. Nature Scientific Reports, 2018.)
- Tools for user behavior analytics e.g. in educational settings (Vanhee et al., Journal of Pathology Informatics, 2019).
Under the hood
Cytomine developers used several tech stacks to build it, They used web-technologies to create the web-based client and make it compatible with the modern web browsers like Google Chrome, Mozilla Firefox, & Safari. They used Java technologies which makes it cross-platform (Linux, macOS, Windows). Here is a list of the tech stack used:
- Java
- Python
- Grails
- Groovy
- OpenSlide: Open source library for processing virtual slides (WSI)
- OpenLayers: open-source JavaScript library for displaying map data in web browsers as slippy maps
- jQuery: HTML DOM manipulation library
- Backbone JS: Functional JavaScript Framework
- PostGIS: Provides spatial objects for the PostgreSQL database
- Hibernate ORM: Object-relational mapping tool for the Java
- OpenCV: Open Computer Vision Library.
- MongoDB: NoSQL DB
- ........ many more.
The project has detailed installation instructions for Linux, & macOS, A demo account can be requested from the project maintainers. It has detailed documentation for developers, and data scientists for scripting, data mining, available algorithms, REST-API, & how to build and add extensions.
License
Cytomine is released as an open-source project under Apache 2.0 license
Resources
- Cytomine's website
- Cytomine's project
- Github
- Cytomine overview (PDF)
Whole-slide Image Analysis
Open source Free Whole-slide Image Viewers and Analysis Software