18 Free and Open-source Whole Slide Imaging Pathology Projects and Libraries, a Comprehensive Guide for Bioengineers and Bio Data Scientists (2024)
Table of Content
Digital pathology is a cutting-edge field that transforms traditional pathology by digitizing glass slides into high-resolution whole slide images (WSIs). These WSIs capture the entire tissue sample on a slide, enabling detailed analysis and diagnostics through digital means.
By using advanced imaging techniques, digital pathology allows pathologists to view, analyze, and share pathology data more efficiently.
WSIs are essential in various applications, including cancer diagnosis, research, education, and telepathology, where they support remote consultations and collaborative studies, improving the accuracy and accessibility of pathological assessments.
Our Digital Pathology Archive



In the following list, we offer you the most active and free digital pathology and WSI (Whole Slide Image) projects in 2024.
1. QuPath
QuPath is an active open-source project for Bioimage analysis & digital pathology.
Features & Tools
- Lots of tools to annotate and view images, including whole slide & microscopy images
- Workflows for brightfield & fluorescence image analysis
- New algorithms for common tasks, including cell segmentation, tissue microarray dearraying
- Interactive machine learning for object & pixel classification
- Customization, batch-processing & data interrogation by scripting
- Easy integration with other tools, including ImageJ
2. CLAM
CLAM (Computational Pathology) is an innovative open-source software developed by the Mahmood Lab. It leverages deep learning models to analyze whole-slide images (WSIs) of histopathological specimens.
CLAM is designed to assist in various aspects of computational pathology, enabling the automatic segmentation, classification, and quantification of tissue regions in WSIs.
This tool is particularly useful in the field of bioinformatics for advancing cancer research and diagnosis by providing a robust and scalable platform for processing large-scale pathology datasets.
3. Tifffile Python Library
Tifffile is a versatile Python library designed for storing and reading images and metadata from various TIFF (Tagged Image File Format) and TIFF-like files commonly used in bioimaging. The library supports a wide range of file formats, including TIFF, BigTIFF, OME-TIFF, GeoTIFF, Adobe DNG, and several specialized formats like Zeiss LSM, ImageJ hyperstack, and more.
Tifffile is particularly useful for researchers working with large-scale image data in bioinformatics and related fields, providing robust tools for both reading and writing complex image file formats.
Features
- Storage and Reading: Tifffile allows for the storage of NumPy arrays in TIFF files and supports reading image data and metadata from numerous TIFF and TIFF-like file formats.
- Supported Formats: The library can handle files such as BigTIFF, OME-TIFF, GeoTIFF, Adobe DNG, and others used in various bioimaging and scientific applications.
- Image Data Handling: It can read image data as NumPy arrays or Zarr arrays/groups from different structures like strips, tiles, pages, SubIFDs, higher-order series, and pyramidal levels.
- Writing Capabilities: Tifffile supports writing image data to TIFF, BigTIFF, OME-TIFF, and ImageJ hyperstack files in various forms, including multi-page, volumetric, pyramidal, and compressed formats.
- Compression Support: The library integrates with imagecodecs to support multiple compression and predictor schemes like LZW, JPEG, JPEG 2000, WebP, and others.
- Advanced Functionality: Tifffile can inspect TIFF structures, handle multi-dimensional file sequences, patch TIFF tag values, and parse proprietary metadata formats, making it a comprehensive tool for managing TIFF files in scientific research.
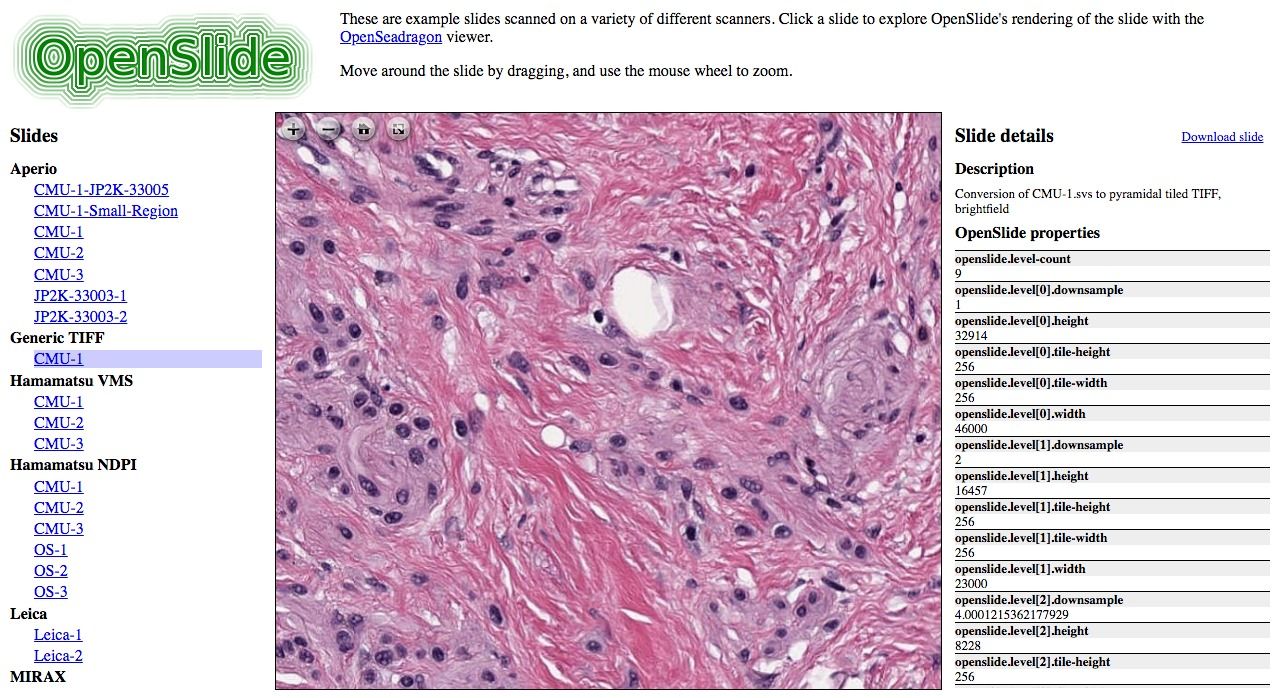
4- OpenSlide
OpenSlide is an open-source C library that facilitates the reading and manipulation of whole-slide images (WSIs), which are high-resolution images of tissue samples used in digital pathology. This library is crucial for bioinformatics and medical imaging research, enabling the efficient handling of large image files produced by digital slide scanners.
OpenSlide can read WSI formats from various scanner vendors, including Aperio, Hamamatsu, Leica, MIRAX, Sakura, Trestle, and Ventana, making it a versatile tool for researchers working
5. Slideflow
Slideflow is an open-source Python library designed to facilitate deep learning workflows with whole-slide images (WSIs) in computational pathology. Developed with flexibility and scalability in mind, Slideflow enables researchers to efficiently manage, preprocess, and analyze large-scale WSI datasets using deep learning models.
Slideflow is particularly valuable for researchers in digital pathology and bioinformatics, offering a comprehensive toolkit for developing and deploying deep learning models on WSI data.
Primary Features
- WSI Management: Slideflow offers tools for managing and processing WSIs, including tissue masking, patch extraction, and image normalization, which are essential steps in preparing data for deep learning.
- Deep Learning Integration: The library is built to integrate seamlessly with popular deep learning frameworks, making it easier to train, evaluate, and deploy models on pathology datasets.
- Dataset Handling: Slideflow provides functionalities for creating and managing large-scale WSI datasets, including support for distributed computing to handle extensive datasets efficiently.
- Flexible Architecture: The library is designed to be adaptable, allowing users to customize and extend it for specific research needs in computational pathology.
Other Features
- Easy-to-use, highly customizable training pipelines
- Robust slide processing and stain normalization toolkit
- Support for training with weakly-supervised or strongly-supervised labels
- Multiple-instance learning (MIL)
- Self-supervised learning (SSL)
- Generative adversarial networks (GANs)
- Explainability tools: Heatmaps, mosaic maps, saliency maps, synthetic histology
- Robust layer activation analysis tools
- Uncertainty quantification
- Interactive user interface for model deployment
6. Bio-Formats
Bio-Formats is a Java library for reading and writing data in life sciences image file formats. It is developed by the Open Microscopy Environment. Bio-Formats is released under the GNU General Public License (GPL); commercial licenses are available from Glencoe Software.
7. DeepSlide
DeepSlide is an open-source Python library designed for deep learning-based analysis of whole-slide images (WSIs) in digital pathology.
The project is developed by the BMIRDS team, DeepSlide provides a streamlined workflow for researchers and clinicians working on computational pathology projects, enabling efficient training, validation, and deployment of deep learning models on WSIs.
DeepSlide is a valuable resource for the computational pathology community, providing a comprehensive toolkit for developing and deploying deep learning models on WSIs. Whether for research or clinical applications, DeepSlide offers the features needed to advance the field of digital pathology.
Features
- Whole-Slide Image Handling: DeepSlide simplifies the processing of WSIs, including patch extraction, tissue segmentation, and image preprocessing, which are essential for accurate deep learning analysis.
- Model Training and Evaluation: The library provides tools to facilitate the training, validation, and testing of deep learning models on pathology image data, making it easier to achieve high-performance results.
- Customizable Pipelines: DeepSlide allows users to create and customize pipelines tailored to specific research needs, offering flexibility in how data is processed and models are trained.
- Integration with Popular Frameworks: DeepSlide is compatible with major deep learning frameworks, such as TensorFlow and PyTorch, enabling seamless integration into existing machine learning workflows.
- Scalability: The library supports distributed computing, making it suitable for handling large datasets and complex analyses in a scalable manner.
- Visualization Tools: DeepSlide includes visualization tools that help researchers interpret model outputs, such as heatmaps and overlay images, to better understand the results of their analysis.
8. DeepLIIF
DeepLIIF is an open-source Python library developed by the Nadeem Lab, designed to enhance and streamline the analysis of immunofluorescence (IF) images in computational pathology.
It leverages deep learning techniques to automate the segmentation, quantification, and analysis of IF images, which are commonly used in biomedical research for visualizing specific proteins, cells, or tissues.
DeepLIIF simplifies the traditionally manual and time-consuming process of analyzing IF images, making it more efficient and reproducible. By providing a robust toolset for researchers, DeepLIIF aids in the accurate interpretation of complex biological data, contributing to advancements in fields like cancer research and cellular biology.
The library's ability to handle large datasets and produce consistent results makes it an essential tool for researchers working with immunofluorescence microscopy.
9. SlideSeg (Python)
This is an open-source (MIT) Python module that produces image patches and annotation masks from whole slide images for deep learning in digital pathology.
10. WSITools
Tools for whole slide image (WSI) processing. Especially for (pairwise) patch extraction, annotation parsing and data preparation for deep learning purposes.
11. WholeSlideData
This is an open-source framework for working with whole-slide images (WSIs) in digital pathology. It includes tools and scripts for handling, processing, and analyzing WSIs, making it easier for researchers to manage large-scale pathology datasets.
The framework is designed for efficient data handling, supporting various formats and enabling streamlined workflows in computational pathology.
12. A graph-transformer for whole slide image classification
The tmi2022 repository by vkola-lab contains the code and resources related to a study published in the IEEE Transactions on Medical Imaging in 2022. The project focuses on developing and evaluating deep learning models for medical image analysis. It provides the necessary scripts, models, and datasets used in the study, allowing for replication and further research in the field of medical imaging.
13. HistomicsStream
The HistomicsStream project is an open-source app by DigitalSlideArchive provides tools for real-time streaming and annotation of whole-slide images (WSIs). It supports live image processing, allowing users to perform annotations and analyses on WSIs as they are being streamed.
This tool is useful for collaborative pathology workflows, enabling multiple users to interact with and analyze large pathology images simultaneously.
14. wsireg
wsireg performs multi-modal or mono-modal whole slide image registration in a graph structure for complex registration tasks using elastix. For detailed introduction and installation and usage instructions see the docs.
Features
- Graph based approach to defining modalities and arbitrary transformation paths between associated images
- Use of
elastix(through ITKElastix) to perform registration - Support for linear and non-linear transformation models
- Transform associated data (masks, shape data) along the same path as the images.
- Supports images converted to OME-TIFF using bioformats2raw -> raw2ometiff pipeline as well as
array_likeimages from memory (np.ndarray,zarr.Array,da.core.Arrayfromnumpy,zarr, anddask, respectively) - All registered images exported as pyramidal OME-TIFF or OME-zarr that can be viewed in software such as Vitessce,vizarr, QuPath, OMERO or any platform that supports these formats.
- All transforms for complex registration paths are internally composited and only 1 interpolation step is performed, avoiding accumulation of interpolation error from many registrations
- Shape data (polygons, point sets, etc.) in GeoJSON format (portable format for QuPath detection/annotation data since v0.3.0) can be imported and transformations applied producing a modified GeoJSON
- Some support for reading native WSI formats: currently reads .czi and .scn but could be expanded to other formats supported by
tifffile
15. WSInfer (Python)
This is an open-source Python-based tool for inferring cell-level information from whole-slide images (WSIs) in digital pathology. It uses machine learning models to analyze WSIs and extract detailed information about cellular structures, aiding in the study and diagnosis of diseases.
The tool is designed to integrate seamlessly with existing workflows in computational pathology, offering a reliable solution for cell-level analysis from high-resolution pathology images.
16. Exact
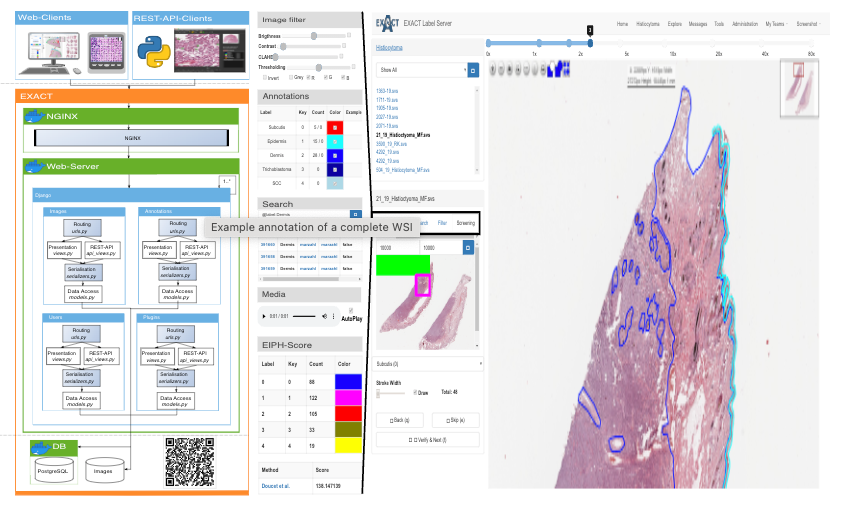
Exact is a free and open source online platform for collaborative image labeling of almost everything.
17. pyslide
The pyslide is a Python library designed for the analysis and processing of whole-slide images (WSIs). It provides tools for extracting, handling, and analyzing regions of interest within WSIs, making it useful for tasks in computational pathology and related research areas.
18- wsic
Whole Slide image (WSI) conversion and compression tool for brightfield histology images.
Features
- Reading and writing several container formats.
- Support for a wide range of compression codecs.
- Custom tile size
- Lossless repackaging / transcoding (to zarr/NGFF or TIFF) from:
- SVS (JPEG compressed)
- OME-TIFF (single image, JPEG and JPEG2000 (J2K) compressed)
- Generic Tiled TIFF (JPEG, JPEG2000, and WebP compressed)
- DICOM WSI (JPEG and JPEG2000 (J2K) compressed)