An open-source Laboratory Information Management System (LIMS) is a system that is developed and distributed with an open-source license, allowing users to access, modify, and distribute the source code.
Advantages and Benefits
Here are five advantages and benefits of using an open-source laboratory system:
- Customizability: Open-source LIMS provides the flexibility to tailor the system to meet the specific needs and workflows of a laboratory. Users can modify the source code and add new features or functionalities according to their requirements, ensuring a customized solution that fits their unique processes.
- Cost-effective: Open-source LIMS eliminates the need for expensive licensing fees associated with proprietary systems. Users can download and use the system for free, significantly reducing the overall cost of implementing and maintaining a laboratory management system.
- Community-driven development: Open-source LIMS projects often have active and diverse communities of developers and users who contribute to the improvement and advancement of the system. This collaborative approach leads to continuous updates, bug fixes, and enhancements, ensuring that the system remains up-to-date and aligned with industry standards.
- Interoperability: Open-source LIMS systems are designed to be interoperable, allowing seamless integration with other laboratory instruments, software, and systems. This interoperability enables data exchange, automation, and streamlined workflows, improving efficiency and reducing manual errors.
- Transparency and security: Open-source LIMS promotes transparency by allowing users to inspect and review the source code. This transparency ensures that the system operates as intended, and any potential security vulnerabilities can be identified and addressed promptly. Additionally, the open-source community actively contributes to the security of the system by conducting code audits and implementing best practices.
Using an open-source laboratory system provides laboratories with the freedom, flexibility, and cost-effectiveness to manage and analyze their data in a way that is tailored to their specific needs.
Here is a list of the best open-source/ free LIMS systems.
1- eLabFTW
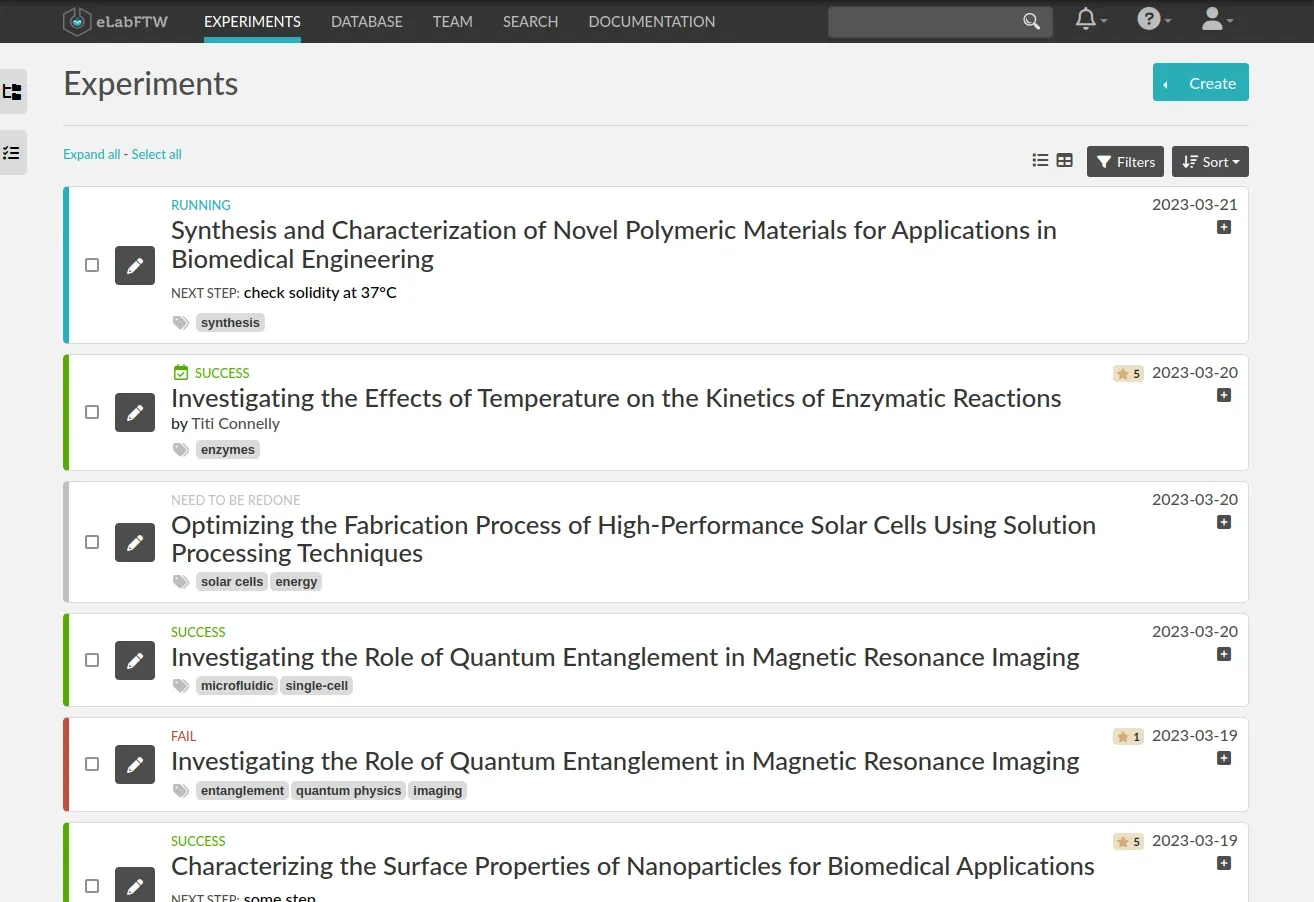
eLabFTW is an electronic lab notebook manager that allows research teams to easily store and organize their experiments.
It features a database for storing various objects and can be accessed via a browser. It can be installed at the institute level to host multiple research teams and is used by research institutions and companies to improve traceability and reproducibility.
Features
Here are some key functionalities of eLabFTW:
- Lab notebook for recording and documenting experiments: With eLabFTW, researchers can maintain a comprehensive lab notebook to keep track of their experimental procedures, observations, and results. This ensures that all crucial details are recorded accurately for future reference.
- Database for managing lab resources: eLabFTW offers a built-in database that enables researchers to efficiently manage various lab resources such as reagents, equipment, storage, cell lines, and more. This centralized repository allows easy access and retrieval of essential information, reducing the chances of errors and duplication.
- Enhanced traceability through trusted and blockchain timestamping: eLabFTW incorporates trusted timestamping mechanisms that provide an additional layer of authenticity to experimental data. Additionally, it offers blockchain timestamping, ensuring the immutability and integrity of the recorded information.
- Import and export functionality in multiple formats: Researchers can seamlessly import and export data in various formats using eLabFTW. This flexibility allows for easy collaboration with other research teams and institutions, promoting knowledge sharing and data exchange.
- Calendar for efficient equipment booking: The integrated calendar feature in eLabFTW enables researchers to manage the booking of lab equipment, ensuring optimal utilization and avoiding conflicts. This streamlines the scheduling process, saving valuable time and resources.
- Support for diverse scientific file formats: eLabFTW supports a wide range of scientific file formats, including text, images, spreadsheets, and more. This versatility enables researchers to work with their preferred file types and ensures compatibility with existing workflows.
- Molecule/equation editor: With the built-in molecule and equation editor, researchers can easily create and edit complex chemical structures and mathematical equations within eLabFTW. This simplifies the documentation of chemical reactions and mathematical models, enhancing the clarity and precision of experimental procedures.
- LaTeX support for scientific writing: eLabFTW offers LaTeX support, allowing researchers to write and format scientific documents with ease. This feature is particularly beneficial for publications, thesis writing, and creating professional-looking reports.
- Todolist for task management: eLabFTW includes a handy todolist feature that helps researchers stay organized and prioritize their tasks. This feature allows users to create, assign, and track tasks, ensuring efficient project management and timely completion of experiments.
- Public REST API for integration: eLabFTW provides a public REST API that enables seamless integration with other research tools and systems. This API allows researchers to customize and automate their workflows, enhancing productivity and data interoperability.
- Multilingual support: eLabFTW is available in 17 languages, making it accessible to research teams from diverse linguistic backgrounds. This multilingual support promotes inclusivity and facilitates global collaboration.
- Advanced permissions system: eLabFTW offers an advanced permissions system that allows administrators to control and manage user access rights. This ensures data privacy and security, protecting sensitive research results from unauthorized access.
- Audited, secure codebase: eLabFTW has undergone rigorous auditing to ensure a secure codebase suitable for handling sensitive research data. This robust security framework provides researchers with peace of mind, knowing that their valuable data is protected.
With its comprehensive set of features and user-friendly interface, eLabFTW has become a trusted choice for research institutions and companies seeking to improve the traceability and reproducibility of their experiments.
2- Pangu LIMS
Pangu LIMS is a powerful and user-friendly web-based Laboratory Information Management System (LIMS) designed to efficiently manage and analyze genomic data.
With Pangu LIMS, researchers can seamlessly handle and interpret genomic data obtained from various wet and dry lab experiments.
This amazing free platform offers a comprehensive set of tools and functionalities to facilitate data organization, quality control, data analysis, and data visualization.
Whether you are conducting DNA sequencing, gene expression analysis, or genotyping experiments, Pangu LIMS provides a centralized and integrated solution for all your genomic data management needs.
3- Viper LIMS
ViperLIMS is an incredibly user-friendly and highly adaptable LIMS system designed specifically for laboratory teams that are constantly evolving and growing. It offers a wide range of features that cater to the unique needs of laboratories.
Features
Some of the key features include:
- Customisable client side: With ViperLIMS, you have the flexibility to build your own VIPER modules using Python and assistance from Boa Constructor. This allows you to tailor the system to your specific requirements and ensure that it meets the needs of your laboratory.
- Boa Constructor: ViperLIMS provides the convenience of Boa Constructor, which enables you to easily create new HTML-Javascript-Python front-end modules through a simple drag and drop interface. This makes it accessible even to users with limited programming experience.
- Client Scripting: Whether you're a beginner or an advanced programmer, ViperLIMS has you covered. It offers a beginner-friendly code structure that allows even those with limited programming knowledge to get started. At the same time, it offers complete control and flexibility for advanced programmers to create more complex scripts.
- Client Commands: With ViperLIMS, you can easily script client to server requests that are passed through Server Methods. This enables seamless communication between the client and server, making it easier to retrieve and manipulate data as needed.
- Server Methods: ViperLIMS provides a comprehensive set of programmable permission-authenticated server-side functions. This allows you to create custom server methods that perform specific actions based on your laboratory's requirements. This level of control ensures that the system operates exactly as you need it to.
- Security: ViperLIMS takes security seriously. It incorporates OAuth2 user verification, which adds an extra layer of protection to the system. Additionally, all HTTP requests made within ViperLIMS are encrypted, ensuring that your data remains secure.
- SQL: ViperLIMS seamlessly integrates with popular database systems such as MySQL, PostgreSQL, and SQLite. This means that you can easily manage and analyze your laboratory data using the database system of your choice.
- OAuth2: ViperLIMS offers intrinsic support for OAuth2 Apps, including popular platforms like Github and LinkedIn. This allows for seamless integration with these platforms, making it easier to authenticate users and access relevant data when needed.
4- Drops LIMS
Drops LIMS is an advanced web application designed specifically for the efficient management of clinical laboratories.
With Drops LIMS, you can streamline and optimize every aspect of your laboratory operations, ensuring smooth and accurate processes from the moment a patient enters your facility to the timely delivery of comprehensive results.
Key Features
Some of the key features offered by Drops LIMS include:
- Streamlined determinations grouped by Biochemical Nomenclator, allowing for efficient and organized tracking of various tests and analyses.
- Comprehensive database of doctors and prescribers, ensuring accurate and up-to-date information for streamlined communication and collaboration.
- Internal worksheets specifically tailored to the needs of your laboratory, providing a seamlessly integrated workflow for your technicians and staff.
- Easy classification of protocols by billing periods, enabling efficient financial management and reporting.
- Practices signed and authorized by qualified biochemical professionals, ensuring compliance with industry standards and regulations.
- Automated generation of detailed PDF reports, which can be easily shared with stakeholders via email for quick and convenient access.
- Real-time monitoring of payments from social works, allowing for effective financial tracking and ensuring timely reimbursement.
- Robust statistical analysis capabilities, providing valuable insights on patient arrival patterns, the most attended social works, patient demographics, billing trends, and more.
- Flexible staff permissions, granting appropriate access and controls to different team members based on their roles and responsibilities.
- Comprehensive system activity log and system logs, ensuring transparency and accountability.
Drops LIMS is not only accessible and user-friendly, but also powerful and feature-rich, providing you with all the necessary tools to enhance efficiency, accuracy, and peace of mind in your laboratory operations.
5- SENAITE.CORE
SENAITE is an exceptional open-source Laboratory Information Management System (LIMS) designed specifically for enterprise environments.
It offers outstanding performance and stability, making it the ideal choice for organizations seeking a reliable and efficient solution for managing their laboratory information.

6- Baobab LIMS
Baobab LIMS is a highly efficient and user-friendly open-source laboratory information management system (LIMS) software. With Baobab LIMS, researchers can easily and accurately track the entire lifecycle of a biospecimen in the laboratory, starting from its initial receipt, throughout storage, and all the way to its eventual reuse.
This comprehensive software ensures that all necessary metadata is meticulously captured, providing researchers with valuable insights and facilitating their work.
Additionally, it is worth noting that Baobab is a common name used to refer to nine different tree species found in various countries around the world, predominantly in Africa.
These unique trees have captured the fascination of people due to their impressive size and distinct appearance.
Baobab LIMS is built using Plone, a highly reliable and robust Python framework that is well-known for its stability and versatility.
Leveraging the power of Plone, Baobab LIMS inherits certain modules from Bika LIMS, further enhancing its functionality and ensuring a seamless user experience.
7- MISO
MISO (Modular and Integrated Sequencing Operations) is a cutting-edge open-source Laboratory Information Management System (LIMS) specifically designed for Next-Generation Sequencing (NGS) sequencing centers.
With its streamlined and user-friendly interface, MISO revolutionizes the way sequencing centers manage their operations, from sample tracking to data analysis and reporting.
One of the key features of MISO is its modular architecture, allowing seamless integration with various NGS platforms and instruments. This flexibility empowers sequencing centers to adapt and scale their workflows according to their specific needs and requirements.
Furthermore, MISO provides comprehensive data management capabilities, ensuring the efficient organization, storage, and retrieval of vast amounts of sequencing data.
This enables researchers and scientists to easily access and analyze their data, accelerating their discoveries and advancing scientific breakthroughs.
8- gP2S
gP2S is a user-friendly web-based laboratory information management system (LIMS) designed for cryo-EM labs and multi-user, multi-project facilities. It tracks various entities and their relationships, allowing for accurate record-keeping.
The system can be used on touchscreen devices and integrates with existing workflows through REST API endpoints.
While currently requiring manual data entry, future versions aim to minimize input by interacting directly with hardware, software, and files. gP2S is still under development and welcomes feedback from users.
9- LIMBuS
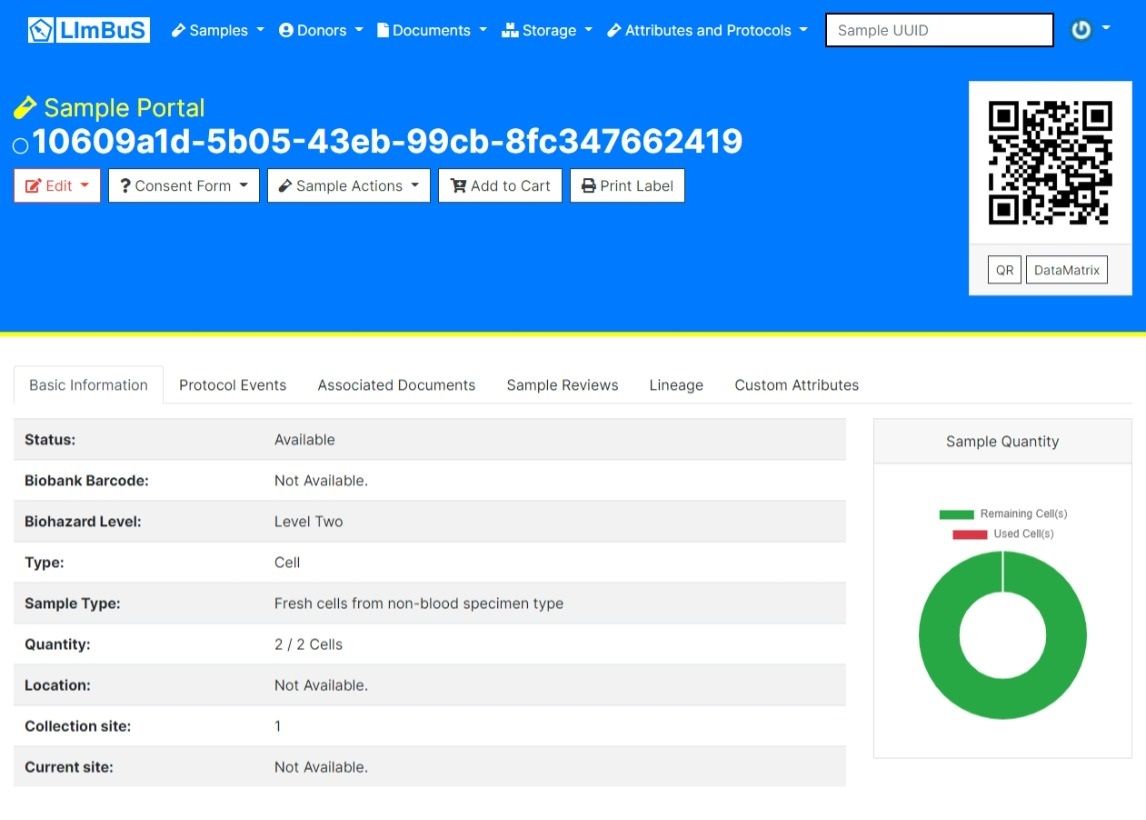
LImBuS, which stands for Laboratory Information Management System, is an exceptional open source and standards compliant Biobank Information Management System.
This innovative system has been developed by a team of dedicated researchers from Aberystwyth University, with the aim of revolutionizing the way biospecimens and associated data are managed.
The primary objective of the LImBuS project is to create a robust and user-friendly Biobank Information Management System (BIMS) that will streamline the entire process of accepting, processing, distributing, and tracking biospecimens. This system will be utilized by the esteemed biorepository at Hywel Dda University Health Board's Clinical Research Centre (CRC), ensuring efficient and accurate management of valuable biological samples.
Features
- Comprehensive Sample Management: LImBuS offers a comprehensive set of features for managing biospecimens, ensuring proper storage, retrieval, and tracking.
- Sample Aliquot and Derivative Creation: With LImBuS, users can easily create aliquots and derivatives from existing samples, allowing for efficient utilization of limited resources.
- Protocol Management: The system provides robust protocol management capabilities, enabling researchers to document and track the specific procedures involved in sample handling and processing.
- Donor Management: LImBuS facilitates efficient donor management, allowing for accurate tracking of donor information, consent, and associated biospecimens.
- Consent Management: The system ensures strict adherence to ethical guidelines by providing a comprehensive consent management module, allowing researchers to document and track consent for each biospecimen.
- Storage Management: LImBuS offers advanced storage management features, allowing users to efficiently allocate, track, and retrieve samples from various storage locations.
- Encrypted Document Management: To ensure data security and confidentiality, LImBuS includes robust encrypted document management capabilities, safeguarding sensitive information.
- Batch Management of Storage Racks: With LImBuS, users can easily manage and track storage racks in batches, streamlining the process of organizing and retrieving samples.
- Clinical Disease Annotation: The system provides an intuitive interface for annotating biospecimens with relevant clinical disease information, enhancing research capabilities.
- Barcode Generation: LImBuS simplifies sample tracking by automatically generating unique barcodes for each biospecimen, ensuring accurate identification and traceability.
- Barcode Label Creation: The system supports the creation of professional barcode labels, facilitating efficient sample identification and organization.
- REST API: LImBuS offers a powerful REST API, allowing for seamless integration with other systems and applications, enhancing interoperability and data exchange.
10- MetaLIMS
MetaLIMS, an open-source laboratory information management system for small metagenomic labs, is no longer actively maintained and may be out of date.
Users are encouraged to use the most recent versions of applications (Ubuntu, MySQL, PHP, Apache) but should be aware that MetaLIMS has not been updated for recent changes. Usage is at the user's own discretion.
11- Lab Manager
Lab Manager is a web-based Laboratory Information System designed to help clinical molecular laboratory managers organize results data quickly and efficiently.
It is written with technologies such as Handlebars, CSS, JavaScript, amCharts, Bootstrap, Font awesome, bcrypt, MySQL, Express, and Session.
The system was developed in collaboration with a real-world laboratory manager, focusing on tracking COVID-19 test results. The data, provided in JSON format, is ingested using JavaScript for easy analysis and display. It is recommended to deploy the system locally due to current simple security measures.
12- Open-LIMS
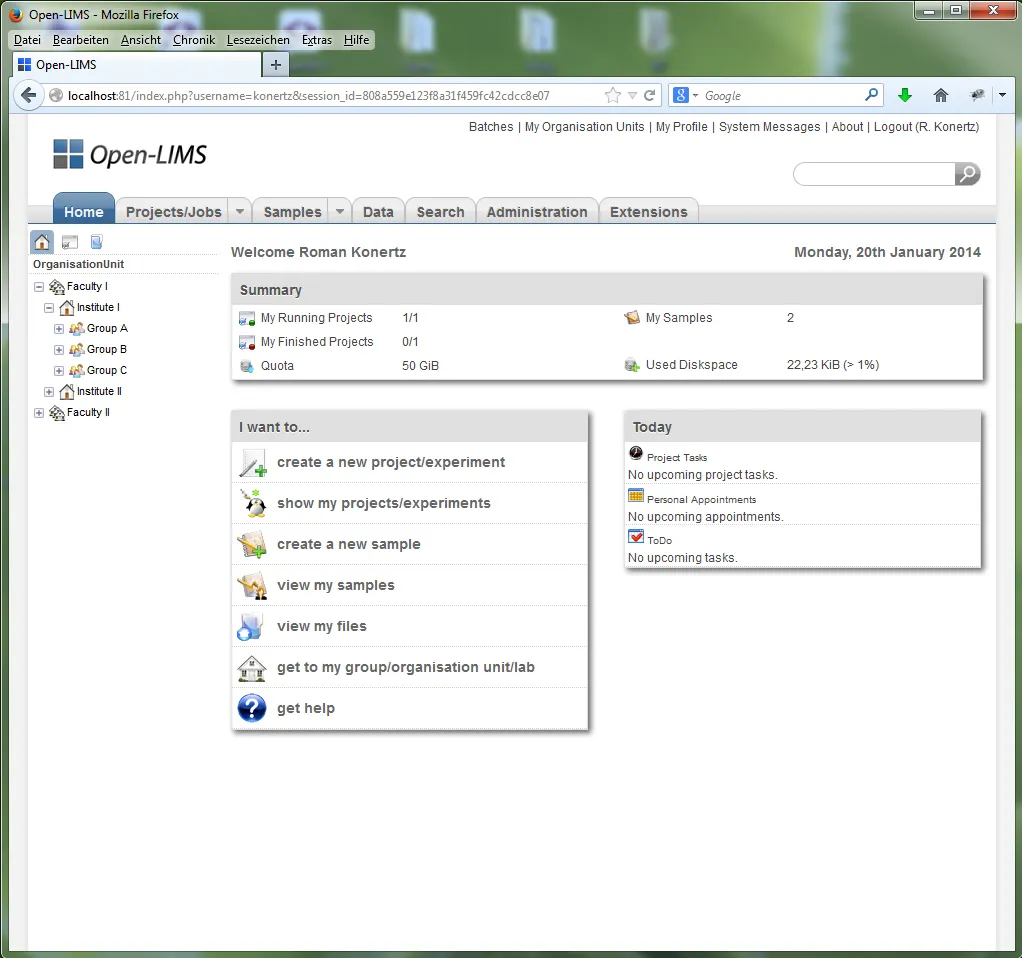
Open-LIMS is a comprehensive project management system for biological projects. It helps with project planning, sample tracking, data management, and collaboration.
With Open-LIMS, researchers can easily organize and track projects, manage samples and data, and streamline workflows. This tool enhances productivity, data accuracy, and scientific discoveries in the field of biology.
13- iSkyLIMS
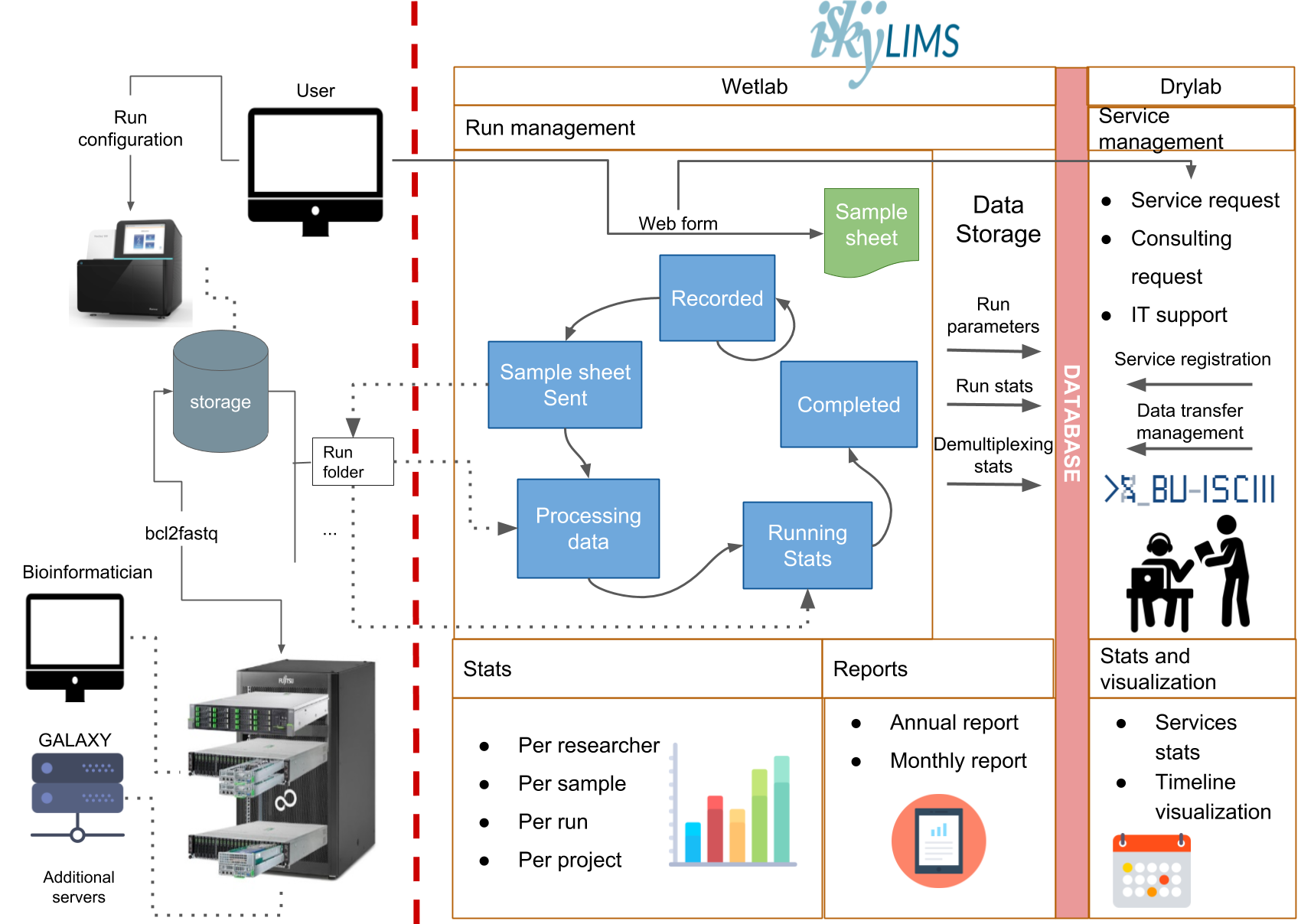
iSkyLIMS is a Laboratory Information Management System (LIMS) designed to handle the tasks involved in genomics facilities, specifically in the context of massive sequencing (MS).
It aims to streamline the workflow from library preparation to data production, reducing errors and facilitating quality control.
Additionally, iSkyLIMS connects the wet lab with the dry lab, enabling easier data analysis by bioinformaticians.
14- NEMO
The NEMO web application is laboratory logistics software created to be easy to use and understand, making lab management simpler. NEMO manages tool reservations, controls tool access, and improves logistics and communication. The code is available for free as open source, enabling other labs to benefit from it.
15- Sequencescape
Sequencescape is a highly versatile and scalable laboratory information management system (LIMS) designed to streamline operations in research labs that handle a large volume of samples.
Benefits
With Sequencescape, labs can benefit from:
- Efficient work order tracking to ensure smooth workflow management.
- Comprehensive sample and study management capabilities for better organization and easy retrieval of data.
- Effective capacity management for pipelines, allowing labs to optimize their resources and prioritize tasks.
- Seamless integration with accounting systems, enabling accurate financial tracking and reporting.
- Accessioning support for samples and studies at the EBI ENA/EGA, ensuring proper identification and documentation.
- Customizable workflows that can be dynamically defined to accommodate the unique processes of each lab.
- Labware and freezer tracking to ensure proper inventory management and sample storage.
- API support for easy integration with third-party applications, enhancing interoperability and data exchange.
The current installation of Sequencescape is already powering the management of over 5 million samples and 1.8 million pieces of labware. It has been successfully adopted by a large organization of 900 people, proving its scalability and effectiveness in meeting the needs of labs with high sample volumes.
16- Aquarium
Aquarium is a laboratory operating system that allows researchers to specify experimental protocols, manage inventory, compute formulae, and present instructions to lab technicians.
It provides a graphical workflow designer and touchscreen monitors for scheduling and logging every step of the experiment.
Aquarium also enables reproducibility by providing an executable description of the results. It is used by the UW BIOFAB at the University of Washington.
17- UniversaLIS
UniversaLIS is a robust and versatile Laboratory Information System (LIS) designed to meet the needs of clinical laboratories. It is specifically developed to be compatible with ASTM/CLSI-compliant clinical laboratory instruments. The system is primarily written in C# programming language, ensuring high performance and reliability.
Originally, UniversaLIS was created to cater to Siemens IMMULITE systems.
However, due to its flexible architecture, it has been expanded to seamlessly integrate with any laboratory system that complies with ASTM-1381/1394 or CLSI-LIS1-A/LIS2-A2 specifications.
With UniversaLIS, clinical laboratories can streamline their operations, enhance efficiency, and improve patient care.
The system offers a wide range of features, including comprehensive instrument management, result reporting, quality control, and data analysis. It provides a user-friendly interface and customizable workflows to adapt to the unique requirements of each laboratory.
18- Biobank
Biobank is an open-source Java application developed by the Canadian BioSample Repository (CBSR) to process and store biospecimens.
It allows multiple users to simultaneously handle specimens, with features for nurses, lab technicians, researchers, and administrators.
Biobank is part of a larger effort to facilitate research collaboration and standardization, and it can be adapted for various storage applications.
Currently, Biobank is used by CBSR to manage a large number of biospecimens and patients across multiple studies and locations.
19- Biobank Web Application
Biobank version 4 is a web-based rewrite of the original application. It follows the principles of Domain Driven Design and uses a CQRS architecture.
Version 4 also includes improvements to the domain model, enhancing workflow and user experience.
For flatbed scanning, a separate desktop client is available. However, this client is only focused on scanning and decoding tubes with 2D DataMatrix barcodes. Not all users need to install the desktop client.
20- labrat
Labrat is a comprehensive framework designed to enhance reproducibility and streamline lab management.
With its wide range of features and functionalities, labrat aims to revolutionize the way scientific experiments are conducted. This package is currently in the initial stages of development, with many exciting updates and improvements on the horizon.
Key Features
Labrat offers a multitude of features that empower researchers and scientists to work more efficiently and effectively. Some key features include:
- Simplified utilization of mathematical functions for diluting solutions, calculating molarity, and more.
- Robust command-line tools for seamless document backup and retrieval.
- Intuitive graphical user interface (GUI) for effortless lab inventory management.
- Streamlined creation and management of new projects using the command-line interface.
Labrat is continuously evolving to meet the ever-changing needs of the scientific community. Stay tuned for upcoming enhancements and exciting additions to this groundbreaking framework.